Abstract
Purpose
Reliable biomarkers for predicting subsequent sepsis among patients with suspected acute infection are lacking. In patients presenting to emergency departments (EDs) with suspected acute infection, we aimed to evaluate the reliability and discriminant ability of 47 leukocyte biomarkers as predictors of sepsis (Sequential Organ Failure Assessment score ≥ 2 at 24 h and/or 72 h following ED presentation).
Methods
In a multi-centre cohort study in four EDs and intensive care units (ICUs), we standardised flow-cytometric leukocyte biomarker measurement and compared patients with suspected acute infection (cohort-1) with two comparator cohorts: ICU patients with established sepsis (cohort-2), and ED patients without infection or systemic inflammation but requiring hospitalization (cohort-3).
Results
Between January 2014 and February 2016, we recruited 272, 59 and 75 patients to cohorts 1, 2, and 3, respectively. Of 47 leukocyte biomarkers, 14 were non-reliable, and 17 did not discriminate between the three cohorts. Discriminant analyses for predicting sepsis within cohort-1 were undertaken for eight neutrophil (cluster of differentiation antigens (CD) CD15; CD24; CD35; CD64; CD312; CD11b; CD274; CD279), seven monocyte (CD35; CD64; CD312; CD11b; HLA-DR; CD274; CD279) and a CD8 T-lymphocyte biomarker (CD279). Individually, only higher neutrophil CD279 [OR 1.78 (95% CI 1.23–2.57); P = 0.002], higher monocyte CD279 [1.32 (1.03–1.70); P = 0.03], and lower monocyte HLA-DR [0.73 (0.55–0.97); P = 0.03] expression were associated with subsequent sepsis. With logistic regression the optimum biomarker combination was increased neutrophil CD24 and neutrophil CD279, and reduced monocyte HLA-DR expression, but no combination had clinically relevant predictive validity.
Conclusions
From a large panel of leukocyte biomarkers, immunosuppression biomarkers were associated with subsequent sepsis in ED patients with suspected acute infection.
Clinical trial registration
NCT02188992.
Similar content being viewed by others
In this first study of standardised multi-site flow cytometry in acutely unwell patients with suspected infections attending emergency departments, we explored which of 47 leukocyte biomarkers reliably discriminates which patients develop sepsis over the next 3 days, defined according to the Sepsis-3 sepsis criteria. |
After highlighting the importance of test reliability (14 biomarkers lacked measurement reliability) and comparator cohorts (a further 17 biomarkers did not discriminate acutely unwell patients with suspected infection from patients with established sepsis-related critical illness and/or non-infective acute illness), we found that none of the remaining 16 biomarkers had clinically relevant predictive ability for subsequent sepsis or other important clinical outcomes. However, markers of early immune suppression (neutrophil and monocyte CD274 and CD279; monocyte HLA-DR) had the strongest associations with clinical outcomes. The optimum biomarker combination associated with clinical deterioration to sepsis was increased neutrophil CD24 and CD279 and reduced monocyte HLA-DR expression. |
Introduction
Sepsis is life-threatening organ dysfunction caused by a dysregulated host response to infection [1]. Host immune responses result from leukocytes sensing pathogen- and tissue damage-associated danger signals [2, 3]. Sepsis-related immune responses involve both humoral and leukocyte components of the innate and adaptive immune systems, with excessive inflammation and immunosuppression occurring simultaneously in most patients [2, 3]. These are thought to influence the resulting clinical phenotypes and outcomes [3, 4].
Leukocyte responses in sepsis measured using flow cytometry detect leukocyte biomarkers, including surface markers and/or leukocyte subsets [5]. Previous flow cytometry-based leukocyte biomarker studies in sepsis were mostly small, single-centre studies in patients with sepsis, typically focusing on a limited panel of biomarkers. These studies rarely evaluated biomarker reliability and reproducibility, which is methodologically and clinically relevant as it influences diagnostic validity [6]. In addition, few studies used robust unbiased designs to assess predictive ability for clinically relevant outcomes in unselected populations with suspected infections prior to developing organ dysfunction and established sepsis.
We hypothesized that among patients with clinically suspected acute infection, but without established sepsis, leukocyte biomarkers would identify patients who subsequently deteriorate clinically and develop sepsis, when measured within a few hours of presentation to the emergency department (ED). Our study objectives were: (1) to identify reliable leukocyte biomarkers; (2) to ascertain which of the reliable biomarkers could discriminate [6] acutely unwell patients with suspected infection from patients with community acquired sepsis-related critical illness in the intensive care unit (ICU) and/or ED patients with non-infective acute illness requiring hospitalisation; and (3) to ascertain whether any of the reliable biomarkers with cross-cohort discrimination could predict which patients with suspected infection in the ED subsequently develop sepsis. We also undertook a post hoc extreme phenotype analysis [7], to compare the biomarker profiles between acutely unwell patients with suspected infection who subsequently developed most severe illness with those who recovered rapidly.
Methods
Study sites and ethics
We performed a prospective, multi-centre, observational cohort study at four sites in the United Kingdom. Ethical approval was granted by the Scotland A/Oxford C Research Ethics Committees (13/SS/0023;13/SC/0266). Consent was provided by patients or surrogate decision-makers according to capacity. We registered the study (NCT02188992) and published the protocol including the analysis plan [8].
Cohort definitions and eligibility criteria
We recruited three distinct patient cohorts using an a priori sampling method to achieve similar age and sex profiles across the ED cohorts. Detailed inclusion/exclusion criteria are listed in the electronic supplement and published protocol (emethods-1) [8]. Cohort-1 comprised acutely unwell patients with suspected infection and systemic inflammation presenting to ED and formed the “discovery cohort”. Patients considered by clinical teams to already have established severe sepsis and/or require ICU admission when screened were excluded. Cohort-2 comprised ICU patients with established community acquired sepsis-related critical illness and formed the “true positive” cohort. Cohort-3 comprised acutely ill patients presenting to ED without infection or systemic inflammation, but requiring hospitalization and formed the “true negative” cohort. Inclusion criteria used throughout the study were based on the sepsis definitions by Levy et al. [9], as our study was designed prior to the Sepsis-3 definitions [1, 10]. All ED patients were enrolled within 12 h of hospital presentation. For all cohorts, we excluded patients with acute pancreatitis, haematological malignancy, chemotherapy in the past 2 weeks, myelodysplastic syndromes, known neutropenia, HIV infection, viral hepatitis infection, pregnancy, blood transfusion > 4 units in the past week, oral corticosteroids for > 24 h prior to enrolment, or a decision not to have active therapy/for palliative care [8].
Leukocyte surface biomarkers and cross-site standardization of flow cytometry
We devised five separate flow cytometry panels to assess 47 leukocyte biomarkers with biological plausibility for having predictive validity for subsequent sepsis (eMethods-1; eTable-1; eFigure-1). We developed, standardized and harmonized flow cytometry procedures across all four study sites [8]. We performed flow cytometry within 4 h of sample acquisition. All anti-human antibodies conjugated to fluorochromes for flow cytometry were from the same batch and clones [all Becton–Dickinson Biosciences (BDB)], standardized on the same platform (FACSCanto II; BDB, San Jose, CA, USA), using a common batch of Cytometer Setup and Tracking beads with the same beads for daily internal quality controls, at all clinical sites. All flow cytometry standard (FCS) files were read by expert technicians using standardized gating procedures developed for each biomarker prior to analysis. The gating strategy for estimating median fluorescence intensity (MFI) or proportions is reported in eMethods-1. All FCS analysis technicians were blinded from clinical data.
Sample size
We based sample size estimates on the confidence interval (CI) widths for positive and negative predictive values (PPV and NPV). The initial design had a primary outcome of septic shock, with an estimated event rate of 5–10% in cohort-1 [11, 12]. For a range in test performance for PPV/NPV of 50–90% we planned a sample size of: cohort-1, n = 300; cohort-2, n = 100; and cohort-3, n = 100, to give a CI width between ± 4.6% to ± 9.8% for PPV and ± 3.4% to ± 6.3% for NPV. At an interim analysis of clinical event rates, the incidence of septic shock was substantially lower than anticipated. We decided by consensus to change the primary outcome to severe sepsis (and subsequently adopted the sepsis-3 sepsis criteria [1] of Sequential Organ Failure Assessment (SOFA) score ≥ 2), with critical care admission a key secondary outcome, to ensure adequate clinically relevant events in the discriminant analyses. These changes occurred prior to study completion and were reported in the published protocol [8].
Statistical analysis
The primary study cohort was cohort-1. The primary exposure was suspected infection. The cohorts-2 and 3 were comparator populations for cross-cohort discrimination and biomarker selection.
Outcomes
The primary outcome was sepsis, defined as SOFA score ≥ 2 at 24 h and/or 72 h following presentation to hospital in patients with suspected infection in the ED (cohort-1) [1]. Secondary outcomes were: critical care admission or death within 72 h of presentation; SOFA ≥ 4 at 24 h and/or 72 h following presentation to hospital; development of septic shock; discharge home within 72 h; discharge to home or in hospital with no organ failure within 72 h; death from sepsis; confirmed infection and length of hospital stay [8]. All cohort-1 data are based on blood samples taken in the ED after recruitment.
Biomarkers selection strategy
Our analytic approach to discover biomarkers with potential diagnostic discrimination for risk of subsequent sepsis occurred in three a priori planned stages and one post hoc analysis.
Stage one: reliability
Inter- and intra-reader reliability for 47 different biomarkers was established according to the protocol [8]. To be included in subsequent evaluation stages, biomarkers needed to demonstrate both inter- and intra-reader reliability at the pre-defined intra-class correlation coefficient (ICC) between readers ≥ 0.9; see Fig. 1 and eMethods-2; eTable-2). For intra-reader reliability the ICC for each reader was calculated as the ratio of within-reader variability to the total variance (within-reader plus residual variance) from the normal linear mixed model. For inter-reader reliability the ICC was calculated as the ratio of between-reader variability to the total variance (between-reader plus residual variance) from the normal linear mixed model. Reliability analyses were done prior to linking leukocyte biomarkers data and clinical outcome data.
Stage two: cross-cohort discrimination
For reliable biomarkers, statistically significant inter-group differences between the three cohorts were explored using Kruskal–Wallis analysis of variance (ANOVA) tests (eTable-3) and visual inspection of data. Biomarkers that discriminated between cohort-1 and either cohort-2 (true-positive) and/or cohort-3 (true negative) and had variability within cohort-1 consistent with potential to discriminate clinical outcomes were selected for Stage-3 analysis. Other factors considered were cell counts, the magnitude of MFI, and potential linkage and co-linearity between groups of biomarkers. This was done in consensus meetings by researchers blinded from clinical outcomes within cohort-1.
Stage three: prediction of clinical outcomes in cohort-1
Within cohort-1 patients, the ability of the selected biomarkers to predict the primary and secondary outcomes was calculated using univariate logistic regression. For the secondary outcomes of death from sepsis, septic shock and length of stay, we provided a descriptive summary as per the analysis plan [8]. The odds ratio (OR) for the outcome per standard deviation increase in biomarker, receiver operating characteristic (ROC) curves, and area under ROC curve (AUROC) were used to assess predictive ability. Youden’s index identified the optimal cut-off point for each marker [13]. Candidate biomarkers that showed consistent inclusion were then taken forward for multivariable modelling.
We used best subsets regression [14] to identify optimal combinations of predictive markers. Specifically, models containing a given number of biomarkers were fitted for all potential biomarker combinations. The five best-fitting models of a given size, according to the Chi squared score statistic, were identified. Biomarkers that consistently appeared in the best-fitting models were selected for the final model. The change in Chi squared score statistic between the best fitting models containing different numbers of biomarkers was used to determine the number of biomarkers to be included in the final model. Linearity of biomarker associations on the logistic scale was checked using plots of deviance residuals. Based on consistency and model fit we identified optimal combinations of predictive markers.
Post hoc extreme phenotype comparison
On the recommendation of a pre-planned independent expert group (see eTable-4), we compared biomarker profiles between sub-populations within cohort-1 with extreme clinical phenotypes of organ dysfunction and outcome to further explore associations for the biomarkers evaluated. We defined well and sick extreme phenotypes [7] by consensus among clinical investigators using clinical data without knowledge of group differences in biomarkers (eFigure-2). The well phenotype had no positive microbiology, a SOFA score ≤ 2 at 24 and 72 h post-enrolment and were either discharged home by 72 h or were in hospital but no longer receiving antibiotics. The sick phenotype had a confirmed infection, SOFA score ≥ 2 at both 24 and 72 h post-enrolment and were still in hospital and receiving antibiotics at 72 h. We compared biomarker expression between the two phenotypes using two-sample t-tests or Mann–Whitney tests as appropriate, applying Bonferroni correction for multiple testing.
For additional comparison, we also measured C-reactive protein (CRP) and procalcitonin (PCT) concentrations at the same time point for cohort-1 patients, given the widespread clinical use of these biomarkers in assessing infection. We constructed ROC curves for CRP and PCT and estimated similar univariate predictive performance characteristics of these for outcomes reported, to enable direct comparison of predictive validity with the more novel biomarkers.
Results
Patient characteristics
Between January 2014 and February 2016, we recruited 272, 59 and 75 patients (N = 406) to cohorts 1, 2, and 3, respectively. The clinical characteristics for the three cohorts and the cohort-1 outcomes are shown in Table 1. Cohorts-1 and 3 had a similar age and sex distribution. Cohort-2 patients tended to be older. The primary outcome in cohort-1, clinical deterioration to sepsis, occurred in 139 patients (51.1%).
Stage one: reliability
The step-wise assessment of intra-reader and then inter-reader reliability resulted in rejection of 14 biomarkers as non-reliable, leaving 33 reliable biomarkers for cross-cohort comparison (Fig. 1; eTable-2).
Stage two: cross-cohort discrimination
Statistical comparison, expert review, and cohort-1 data distribution resulted in rejection of a further 17 biomarkers (Fig. 1; eTable-2; eTable-3). The cross-cohort comparisons plots for the 16 selected biomarkers are shown in eFigure-3. Based on the stage-1 and -2 selections, eight neutrophil biomarkers [cluster of differentiation antigens (CD) CD15; CD24; CD35; CD64; CD312; CD11b; CD274; CD279], seven monocyte biomarkers (CD35; CD64; CD312; CD11b; HLA-DR; CD274; CD279) and one CD8 T-lymphocyte biomarker (CD279) were selected for evaluation of discrimination for clinical outcomes. Biological relevance of these markers in sepsis are summarized in Table 2.
Stage three: prediction of clinical outcomes in cohort-1
Most biomarkers lacked any clinically or statistically significant discrimination for predicting primary and secondary outcomes within cohort-1 patients. Amongst the individual biomarkers, clinical deterioration to sepsis was associated with higher neutrophil CD279 expression, higher monocyte CD279 expression and lower monocyte HLA-DR expression. The optimal MFI cutoff for neutrophil CD279 was 239 [sensitivity 0.88 (95% confidence interval 0.82–0.93); specificity 0.35(0.26–0.43)]; for monocyte CD279 was 141 [sensitivity 0.83(0.77–0.90); specificity 0.39(0.30–0.47)]; and for monocyte HLA-DR was 3572 [sensitivity 0.43(0.34–0.51); specificity 0.69(0.60–0.77)]. Although these associations were statistically significant, discriminant ability was poor and unlikely to be clinically useful in isolation.
With best subsets logistic regression, the optimum combination for predicting clinical deterioration to sepsis included increased neutrophil CD24; increased neutrophil CD279; and reduced monocyte HLA-DR expression [sensitivity 0.72(0.64–0.79); specificity 0.56(0.48–0.65)]. With best subsets logistic regression, the optimum combination for predicting the secondary outcome of discharge to home within 72 h, included increased neutrophil CD15, reduced neutrophil CD274 and increased total monocyte HLA-DR expression. No biomarkers had significant discriminant value for the outcome of critical care admission or death within 72 h. The performance of individual and optimized combinations of biomarkers for predicting the primary and secondary outcomes are shown in Table 3. No marked non-linearities in biomarker effects were identified. Overall, although statistically significant associations were demonstrated, discrimination of clinical outcomes was unlikely to be clinically useful (Fig. 1).
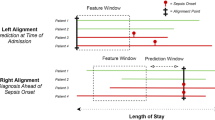
Overview of selection of leukocyte biomarkers for discriminant analysis through the pre-defined stages of the study. For a detailed description of the rationale for biomarker selection see eMethods-1 and eMethods-2. Non-reliable refers to the analysis of cell populations that are not sufficiently distinct in bimodal FACS plots, are difficult to reliably standardize for a uniform gating approach and need further development. We are proposing that these biomarkers are necessarily of limited value
Extreme phenotype analysis
From 272 patients in cohort-1, we identified 40 “well' and 52 “sick” phenotypes (eFigure-2). “Sick” phenotype patients were characterized by being older, more often male, with a higher frequency of co-morbidities, more frequently lymphopenic, with higher APACHE II and SOFA scores at baseline. After Bonferroni correction for multiple comparisons, “sick” phenotypes had significantly higher monocyte CD279 and neutrophil CD279 in the ED, but no other biomarkers were different (Table 4; eFigure-4).
For both CRP and PCT, there was also no statistically or clinically significant discrimination for subsequent sepsis with univariate analysis (Table 3).
Discussion
In this multi-site cohort study, we reduced a candidate panel of 47 leukocyte biomarkers to 16 reliable biomarkers with potential for discriminating the risk of developing sepsis in patients with suspected infection presenting to the ED. The combination of higher neutrophil CD24, higher neutrophil CD279, and a lower monocyte HLA-DR expression best predicted the clinical deterioration to sepsis. Consistent with this association, a lower neutrophil CD279 expression and higher monocyte HLA-DR expression were associated with discharge home within 72 h (implying rapid recovery). Although our pre-defined biomarker discovery strategy identified these biomarkers as associated with development of sepsis and more severe illness, their discriminant value was insufficient to suggest utility for decision-making in routine clinical care.
Our findings have potential clinical relevance. The key pathophysiological insight is that leukocyte biomarkers of immunosuppression such as check-point inhibitors (CD279; CD274) and antigen processing ability (HLA-DR) were altered even in patients with suspected infection presenting to ED. We also demonstrate the importance of assessing reliability when standardising flow cytometry for large-scale time critical use. The development of clinically useable tests is likely to require a form of cross-platform calibration (such as multiparametric version of the Quantibrite system, BD Bioscience). Our study shows it is feasible to implement flow cytometry as a means of undertaking precision medicine in sepsis, for example to guide novel therapeutic interventions such as those tested recently in immunotherapy trials [15] and highlighted in recent expert reviews [16, 17]. However, our data suggest that for patients with suspected infection the predictive validity of panels of leukocyte biomarkers are unlikely to be useful as general clinical decision-making tools. Of note, both CRP and PCT also performed poorly.
Strengths of our study were well-defined hypothesis, pre-published protocol [8], internationally accepted primary outcome [1], clinically relevant secondary outcomes and hierarchical analytic approach to reduce biomarker selection bias. Reliability of multi-site flow cytometry is potentially problematic due to measurement error bias [18], which we addressed rigorously with fluorochrome-conjugated antibody titrated for optimal signal and kept constant throughout the study. Using hospitalized non-infected patients and ICU-sepsis patients as comparators during biomarker selection increased the chance of detecting infection related host responses and is superior to using healthy volunteer controls. Our blood sampling time point in the ED was prior to severe illness, before major clinical interventions, and much earlier than in previous studies of sepsis biomarkers, and we excluded patients who clinicians considered to have already established sepsis and/or critical illness. As such, our population was different from other recent studies, which evaluated leukocyte biomarkers for prediction of sepsis trajectory (by including patients with sepsis-2 defined sepsis, severe sepsis and septic shock) [19, 20] and stratified nosocomial infection risk in ICU patients [21] (see eTable-5, which highlights important differences). The post hoc extreme phenotype analysis enhanced face validity by considering multiple clinical variables simultaneously for phenotype definition.
Our study has potential weaknesses. Although we could not include all potential leukocyte biomarkers, we studied a range of leukocyte biomarkers (such as complement pathway receptors (CD35, CD11b), G protein-coupled receptors (CD312), Fc-gamma-receptors (CD64 [22, 23]), factors delaying neutrophil apoptosis (CD24 [22]), check-point molecules (CD274, CD279) [24]; HLA-DR expression [25,26,27]), that previous studies highlight association with adverse outcomes in established sepsis. We enrolled a smaller sample size than planned due to time and funding constraints. However, this had a limited impact since substantial differences in biomarker levels across cohorts still enabled selection of candidate biomarkers for further discriminant analysis. Supervised classification methods such as classification and regression trees (CART) is a valid alternative analytic approach for this research question. However, CART requires approximately 50 events per variable when predicting a dichotomised outcome, before predictions become stable and over-optimism is minimised [28]. As our observed number of sepsis events did not reach this threshold we opted to use the best subsets logistic regression approach as pre-specified in our statistical analysis plan [8]. As our cohort-1 inclusion criteria mandated SIRS, we have excluded SIRS negative patients with infection, who could have progressed to develop sepsis. However, this is unlikely to bias the results, as the prevalence of SIRS negative sepsis-3 sepsis in ICUs in England is only 3% [29]. As our objective was to study leukocyte biomarkers at an earlier time point than previously achieved and to identify biomarkers that predict deterioration within 72 h of hospitalisation, we excluded patients planned for direct admission to ICU from the ED at enrolment, which explains the lower than expected event rate for death and septic shock. Findings might be different for more severely ill patients studied later in sepsis, as observed in other recent flowcyometric studies (eTable-5) [19,20,21].
Our findings have biological plausibility, as the leukocyte biomarkers that best predicted the risk of developing sepsis in our study were on the key innate immune cells, namely neutrophils and monocytes, which are first responders to infection. The strongest biomarker predicting subsequent sepsis and extreme phenotypes was higher levels of CD279 (programmed death receptor 1, PD-1) on monocytes and neutrophils. CD279 expression is associated with neutrophil and monocyte suppressor subsets [30], memory lymphocyte subsets [31], is thought to regulate T cell responses and induce an inhibitory signal characterized by cell cycle arrest and reduced cytokine synthesis [2, 32]. This early role for CD279/PD-1 is consistent with animal models of sepsis [33] and sepsis cohorts [30]. CD279/PD-1 acts in conjunction with its ligand CD274 (PD-L1). In our study, lower CD274, together with lower CD279, higher monocyte HLA-DR, and lower neutrophil CD24, emerged as a predictor for rapid recovery sepsis phenotype. These novel findings require further confirmatory studies.
Although none of the biomarkers we studied had discriminant ability that could be used to guide clinical decision-making, our data imply that immunosuppression in infected patients precedes established sepsis and that higher CD279/PD-1 and lower HLA-DR are potential theragnostic and enrichment markers [34,35,36,37] for anti-PD-1/PDL-1 agents and granulocyte-monocyte colony stimulating factor [25], respectively, for carefully designed immunotherapy trials [3, 38].
Conclusions
We conclude that in a population of patients presenting with suspected infection prior to established sepsis, a sequential approach to identifying reliable potential leukocyte biomarkers from a large candidate panel that may predict the subsequent development of sepsis identified only a small number with discriminant properties. These were markers of immune suppression, namely CD279 and HLA-DR, suggesting this may be an early event, prior to development of sepsis.
References
Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche JD, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent JL, Angus DC (2016) The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 315:801–810
Hotchkiss RS, Monneret G, Payen D (2013) Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol 13:862–874
van der Poll T, van de Veerdonk FL, Scicluna BP, Netea MG (2017) The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol 17:407–420
Scicluna BP, van Vught LA, Zwinderman AH, Wiewel MA, Davenport EE, Burnham KL, Nurnberg P, Schultz MJ, Horn J, Cremer OL, Bonten MJ, Hinds CJ, Wong HR, Knight JC, van der Poll T, consortium M (2017) Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir Med 5:816–826
Venet F, Guignant C, Monneret G (2011) Flow cytometry developments and perspectives in clinical studies: examples in ICU patients. Methods Mol Biol 761:261–275
Angus DC, Seymour CW, Coopersmith CM, Deutschman CS, Klompas M, Levy MM, Martin GS, Osborn TM, Rhee C, Watson RS (2016) A framework for the development and interpretation of different sepsis definitions and clinical criteria. Crit Care Med 44:e113–e121
Shankar-Hari M (2017) How could we enhance translation of sepsis immunology to inform immunomodulation trials in sepsis? Crit Care 21:125
Datta D, Conway Morris A, Antonelli J, Warner N, Brown KA, Wright J, Simpson AJ, Rennie J, Hulme G, Lewis SM, Mare TA, Cookson S, Weir CJ, Dimmick I, Keenan J, Rossi AG, Shankar-Hari M, Walsh TS, Ex PSI (2016) Early PREdiction of Severe Sepsis (ExPRES-Sepsis) study: protocol for an observational derivation study to discover potential leucocyte cell surface biomarkers. BMJ Open 6:e011335
Levy MM, Fink MP, Marshall JC, Abraham E, Angus D, Cook D, Cohen J, Opal SM, Vincent JL, Ramsay G, Sccm/Esicm/Accp/Ats/Sis (2003) 2001 SCCM/ESICM/ACCP/ATS/SIS international sepsis definitions conference. Crit Care Med 31:1250–1256
Shankar-Hari M, Phillips GS, Levy ML, Seymour CW, Liu VX, Deutschman CS, Angus DC, Rubenfeld GD, Singer M, Sepsis Definitions Task F (2016) Developing a new definition and assessing new clinical criteria for septic shock: for the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 315:775–787
Gray A, Ward K, Lees F, Dewar C, Dickie S, McGuffie C, committee Ss (2013) The epidemiology of adults with severe sepsis and septic shock in Scottish emergency departments. Emerg Med J 30:397–401
Glickman SW, Cairns CB, Otero RM, Woods CW, Tsalik EL, Langley RJ, van Velkinburgh JC, Park LP, Glickman LT, Fowler VG Jr, Kingsmore SF, Rivers EP (2010) Disease progression in hemodynamically stable patients presenting to the emergency department with sepsis. Acad Emerg Med 17:383–390
Fluss R, Faraggi D, Reiser B (2005) Estimation of the Youden Index and its associated cutoff point. Biometrical journal Biometrische Zeitschrift 47:458–472
Miller A, Tibshirani R, Cox D, Keiding N, Isham V, Louis T, Tong H, Reid N (2002) Subset selection in regression. Chapman and Hall/CRC, New York
Francois B, Jeannet R, Daix T, Walton AH, Shotwell MS, Unsinger J, Monneret G, Rimmele T, Blood T, Morre M, Gregoire A, Mayo GA, Blood J, Durum SK, Sherwood ER, Hotchkiss RS (2018) Interleukin-7 restores lymphocytes in septic shock: the IRIS-7 randomized clinical trial. JCI insight. https://doi.org/10.1172/jci.insight.98960
Perner A, Gordon AC, Angus DC, Lamontagne F, Machado F, Russell JA, Timsit JF, Marshall JC, Myburgh J, Shankar-Hari M, Singer M (2017) The intensive care medicine research agenda on septic shock. Intensive Care Med 43:1294–1305
Perner A, Rhodes A, Venkatesh B, Angus DC, Martin-Loeches I, Preiser JC, Vincent JL, Marshall J, Reinhart K, Joannidis M, Opal SM (2017) Sepsis: frontiers in supportive care, organisation and research. Intensive Care Med 43:496–508
Mittag A, Tarnok A (2009) Basics of standardization and calibration in cytometry—a review. J Biophotonics 2:470–481
Daix T, Guerin E, Tavernier E, Mercier E, Gissot V, Herault O, Mira JP, Dumas F, Chapuis N, Guitton C, Bene MC, Quenot JP, Tissier C, Guy J, Piton G, Roggy A, Muller G, Legac E, de Prost N, Khellaf M, Wagner-Ballon O, Coudroy R, Dindinaud E, Uhel F, Roussel M, Lafon T, Jeannet R, Vargas F, Fleureau C, Roux M, Allou K, Vignon P, Feuillard J, Francois B, Septiflux Trial G (2018) Multicentric Standardized Flow Cytometry Routine Assessment of Patients With Sepsis to Predict Clinical Worsening. Chest 154:617–627
Guerin E, Orabona M, Raquil MA, Giraudeau B, Bellier R, Gibot S, Bene MC, Lacombe F, Droin N, Solary E, Vignon P, Feuillard J, Francois B (2014) Circulating immature granulocytes with T-cell killing functions predict sepsis deterioration. Crit Care Med 42:2007–2018
Conway Morris A, Datta D, Shankar-Hari M, Stephen J, Weir CJ, Rennie J, Antonelli J, Bateman A, Warner N, Judge K, Keenan J, Wang A, Burpee T, Brown KA, Lewis SM, Mare T, Roy AI, Hulme G, Dimmick I, Rossi AG, Simpson AJ, Walsh TS (2018) Cell-surface signatures of immune dysfunction risk-stratify critically ill patients: INFECT study. Intensive Care Med 44:627–635
Parlato M, Souza-Fonseca-Guimaraes F, Philippart F, Misset B, Captain Study G, Adib-Conquy M, Cavaillon JM (2014) CD24-triggered caspase-dependent apoptosis via mitochondrial membrane depolarization and reactive oxygen species production of human neutrophils is impaired in sepsis. J Immunol 192:2449–2459
Gros A, Roussel M, Sauvadet E, Gacouin A, Marque S, Chimot L, Lavoue S, Camus C, Fest T, Le Tulzo Y (2012) The sensitivity of neutrophil CD64 expression as a biomarker of bacterial infection is low in critically ill patients. Intensive Care Med 38:445–452
Chang K, Svabek C, Vazquez-Guillamet C, Sato B, Rasche D, Wilson S, Robbins P, Ulbrandt N, Suzich J, Green J, Patera AC, Blair W, Krishnan S, Hotchkiss R (2014) Targeting the programmed cell death 1: programmed cell death ligand 1 pathway reverses T cell exhaustion in patients with sepsis. Crit Care 18:R3
Meisel C, Schefold JC, Pschowski R, Baumann T, Hetzger K, Gregor J, Weber-Carstens S, Hasper D, Keh D, Zuckermann H, Reinke P, Volk HD (2009) Granulocyte-macrophage colony-stimulating factor to reverse sepsis-associated immunosuppression: a double-blind, randomized, placebo-controlled multicenter trial. Am J Respir Crit Care Med 180:640–648
Rangel-Frausto MS, Pittet D, Costigan M, Hwang T, Davis CS, Wenzel RP (1995) The natural history of the systemic inflammatory response syndrome (SIRS). A prospective study. JAMA 273:117–123
Rangel-Frausto MS, Pittet D, Hwang T, Woolson RF, Wenzel RP (1998) The dynamics of disease progression in sepsis: Markov modeling describing the natural history and the likely impact of effective antisepsis agents. Clin Infect Dis 27:185–190
van der Ploeg T, Austin PC, Steyerberg EW (2014) Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med Res Methodol 14:137
Shankar-Hari M, Harrison DA, Rowan KM (2016) Differences in impact of definitional elements on mortality precludes international comparisons of sepsis epidemiology—a Cohort Study illustrating the need for standardized reporting. Crit Care Med 44:2223–2230
Patera AC, Drewry AM, Chang K, Beiter ER, Osborne D, Hotchkiss RS (2016) Frontline Science: defects in immune function in patients with sepsis are associated with PD-1 or PD-L1 expression and can be restored by antibodies targeting PD-1 or PD-L1. J Leukoc Biol 100:1239–1254
Wilson JK, Zhao Y, Singer M, Spencer J, Shankar-Hari M (2018) Lymphocyte subset expression and serum concentrations of PD-1/PD-L1 in sepsis—pilot study. Crit Care 22:95
Boomer JS, To K, Chang KC, Takasu O, Osborne DF, Walton AH, Bricker TL, Jarman SD 2nd, Kreisel D, Krupnick AS, Srivastava A, Swanson PE, Green JM, Hotchkiss RS (2011) Immunosuppression in patients who die of sepsis and multiple organ failure. JAMA 306:2594–2605
Wang JF, Li JB, Zhao YJ, Yi WJ, Bian JJ, Wan XJ, Zhu KM, Deng XM (2015) Up-regulation of programmed cell death 1 ligand 1 on neutrophils may be involved in sepsis-induced immunosuppression: an animal study and a prospective case-control study. Anesthesiology 122:852–863
Pene F, Courtine E, Cariou A, Mira JP (2009) Toward theragnostics. Crit Care Med 37:S50–S58
Shankar-Hari M, Rubenfeld GD (2017) The use of enrichment to reduce statistically indeterminate or negative trials in critical care. Anaesthesia 72:560–565
Prescott HC, Calfee CS, Thompson BT, Angus DC, Liu VX (2016) Toward smarter lumping and smarter splitting: rethinking strategies for sepsis and acute respiratory distress syndrome clinical trial design. Am J Respir Crit Care Med 194:147–155
Hotchkiss RS, Sherwood ER (2015) Immunology. Getting sepsis therapy right. Science 347:1201–1202
Venet F, Monneret G (2018) Advances in the understanding and treatment of sepsis-induced immunosuppression. Nat Rev Nephrol 14:121–137
Jalava-Karvinen P, Hohenthal U, Laitinen I, Kotilainen P, Rajamaki A, Nikoskelainen J, Lilius EM, Nuutila J (2009) Simultaneous quantitative analysis of Fc gamma RI (CD64) and CR1 (CD35) on neutrophils in distinguishing between bacterial infections, viral infections, and inflammatory diseases. Clin Immunol 133:314–323
Wang X, Li ZY, Zeng L, Zhang AQ, Pan W, Gu W, Jiang JX (2015) Neutrophil CD64 expression as a diagnostic marker for sepsis in adult patients: a meta-analysis. Crit Care 19:245
Lewis SM, Treacher DF, Edgeworth J, Mahalingam G, Brown CS, Mare TA, Stacey M, Beale R, Brown KA (2015) Expression of CD11c and EMR2 on neutrophils: potential diagnostic biomarkers for sepsis and systemic inflammation. Clin Exp Immunol 182:184–194
Netea MG, Balkwill F, Chonchol M, Cominelli F, Donath MY, Giamarellos-Bourboulis EJ, Golenbock D, Gresnigt MS, Heneka MT, Hoffman HM, Hotchkiss R, Joosten LAB, Kastner DL, Korte M, Latz E, Libby P, Mandrup-Poulsen T, Mantovani A, Mills KHG, Nowak KL, O’Neill LA, Pickkers P, van der Poll T, Ridker PM, Schalkwijk J, Schwartz DA, Siegmund B, Steer CJ, Tilg H, van der Meer JWM, van de Veerdonk FL, Dinarello CA (2017) A guiding map for inflammation. Nat Immunol 18:826–831
Pillay J, Kamp VM, van Hoffen E, Visser T, Tak T, Lammers JW, Ulfman LH, Leenen LP, Pickkers P, Koenderman L (2012) A subset of neutrophils in human systemic inflammation inhibits T cell responses through Mac-1. J Clin Invest 122:327–336
Pillay J, Tak T, Kamp VM, Koenderman L (2013) Immune suppression by neutrophils and granulocytic myeloid-derived suppressor cells: similarities and differences. Cell Mol Life Sci 70:3813–3827
Mare TA, Treacher DF, Shankar-Hari M, Beale R, Lewis SM, Chambers DJ, Brown KA (2015) The diagnostic and prognostic significance of monitoring blood levels of immature neutrophils in patients with systemic inflammation. Crit Care 19:57
Jamsa J, Huotari V, Savolainen ER, Syrjala H, Ala-Kokko T (2015) Kinetics of leukocyte CD11b and CD64 expression in severe sepsis and non-infectious critical care patients. Acta Anaesthesiol Scand 59:881–891
Brunialti MK, Martins PS, Barbosa de Carvalho H, Machado FR, Barbosa LM, Salomao R (2006) TLR2, TLR4, CD14, CD11B, and CD11C expressions on monocytes surface and cytokine production in patients with sepsis, severe sepsis, and septic shock. Shock 25:351–357
Wagner C, Kotsougiani D, Pioch M, Prior B, Wentzensen A, Hansch GM (2008) T lymphocytes in acute bacterial infection: increased prevalence of CD11b(+) cells in the peripheral blood and recruitment to the infected site. Immunology 125:503–509
Demaret J, Venet F, Friggeri A, Cazalis MA, Plassais J, Jallades L, Malcus C, Poitevin-Later F, Textoris J, Lepape A, Monneret G (2015) Marked alterations of neutrophil functions during sepsis-induced immunosuppression. J Leukoc Biol 98:1081–1090
Acknowledgements
This independent research by Dr. Manu Shankar-Hari is supported by the National Institute for Health Research Clinician Scientist Award (NIHR-CS-2016-16-011). The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research or the Department of Health. Prof Christopher Weir was supported in this work by NHS Lothian via the Edinburgh Clinical Trials Unit. Dr. Andrew Conway Morris is supported by a Clinical Research Career Development Fellowship from the Wellcome Trust (WT 2055214/Z/16/Z).
Funding
The study was funded by Innovate UK (Sepsis 2: 101193). Dr Shankar-Hari is supported by the National Institute for Health Research Clinician Scientist Award (CS-2016-16-011). Dr Conway Morris is supported by a Clinical Research Career Development Fellowship from the Welcome Trust (WT 2055214/Z/16/Z).
Author information
Authors and Affiliations
Contributions
Drs. Shankar-Hari, Weir and Walsh had full access to all the data in the study and take responsibility for integrity of data and the accuracy of the data analyses. Concept and design: Walsh, Simpson, Conway Morris, Datta, Weir, Warner. Statistical analysis: Assi, Stephen, Weir, Datta, Wilson, Shankar-Hari. Drafting of manuscript: Shankar-Hari, Weir, Walsh. Acquisition, analysis and interpretation of data: All authors. Critical revision of the manuscript for important intellectual content: All authors. Obtained funding: Walsh, Conway Morris, Brown, Simpson, Warner, Keenan. Administrative, technical, or material support: Walsh, Weir, Warner, Judge. Keenan. Supervision: Walsh, Weir. All authors confirm to the accuracy or integrity of the work.
Corresponding author
Ethics declarations
Conflicts of interests
Noel Warner, Kevin Judge, Jim Keenen and Alice Wang were all employees of BD biosciences whilst this work was being undertaken, and all four authors hold stock in BD Biosciences. Prof Simpson collaborated with BDB on a Wellcome Trust/Department of Health-funded Healthcare Innovation Challenge Fund (HICF) grant in suspected ventilator-associated pneumonia. He is Director of the NIHR Newcastle In Vitro Diagnostic Evidence Co-operative (formerly the NIHR Newcastle Diagnostic Evidence Co-operative)—these entities exist to evaluate in vitro diagnostics and have worked with (and continue to work with) BDB and other companies in this capacity. All other authors declare that they do have any personal conflict of interest directly related to this manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shankar-Hari, M., Datta, D., Wilson, J. et al. Early PREdiction of sepsis using leukocyte surface biomarkers: the ExPRES-sepsis cohort study. Intensive Care Med 44, 1836–1848 (2018). https://doi.org/10.1007/s00134-018-5389-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00134-018-5389-0