Abstract
A minimally invasive and repeatable approach for real-time epidermal growth factor receptor (EGFR) mutation surveillance would be highly beneficial for individualized therapy of late stage lung cancer patients whose surgical specimens are often not available. We aim to develop a viable method to detect EGFR mutations in single circulating tumor cells (CTCs). Using a model CTC system of spiked tumor cells in whole blood, we evaluated EGFR mutation determination in single tumor cells enriched from blood. We used magnetic beads labeled with antibody against leukocyte surface antigens to deplete leukocytes and enrich native CTCs independent of epithelial marker expression level. We then used laser cell microdissection (LCM) to isolate individual CTCs, followed by whole-genome amplification of the DNA for exon 19 microdeletion, L858R and T790M mutation detection by PCR sequencing. EGFR mutations were successfully measured in individual spiked tumor cells enriched from 7.5 ml whole blood. Whole-genome amplification provided sufficient DNA for mutation determination at multiple sites. Ninety-five percent of the single CTCs microdissected by LCM (19/20) yielded PCR amplicons for at least one of the three mutation sites. The amplification success rates were 55 % (11/20) for exon 19 deletion, 45 % (9/20) for T790M, and 85 % (17/20) for L858R. Sequencing of the amplicons showed allele dropout in the amplification reactions, but mutations were correctly identified in 80 % of the amplicons. EGFR mutation determination from single captured tumor cells from blood is feasible with the approach described here. However, to overcome allele dropout and to obtain reliable information about the tumor's EGFR status, multiple individual tumor cells should be assayed.

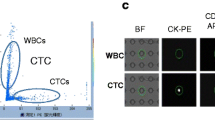
Captured CTC from patient blood. CTC (green arrow) and WBCs (red arrow) enriched from 7.5 ml peripheral blood of a NSCLC patient were immunostained with anti-Cytokeratin 18 (green) and anti-CD45 (red) antibodies. Cell nuclei were stained with DAPI (blue). Compared with H1975 (Fig. 1), CTC from the NSCLC patient appeared to have reduced CK18 expression





Similar content being viewed by others
Abbreviations
- CTC:
-
Circulating tumor cells
- EGFR :
-
Epidermal growth factor receptor
- LCM:
-
Laser capture microdissection
- NSCLC:
-
Non-small cell lung cancer
- TKI:
-
Tyrosine kinase inhibitor
- WGA:
-
Whole-genome amplification
References
Lynch TJ, Bell DW, Sordella R et al (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350:2129–2139
Paez JG, Janne PA, Lee JC et al (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304:1497–1500
Costa DB, Kobayashi S, Tenen DG et al (2007) Pooled analysis of the prospective trials of gefitinib monotherapy for EGFR-mutant non-small cell lung cancers. Lung Cancer 58:95–103
Horiike A, Kimura H, Nishio K et al (2007) Detection of epidermal growth factor receptor mutation in transbronchial needle aspirates of non-small cell lung cancer. Chest 131:1628–1634
Allard WJ, Matera J, Miller MC et al (2004) Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 10:6897–6904
Punnoose EA, Atwal SK, Spoerke JM et al (2010) Molecular biomarker analyses using circulating tumor cells. PLoS One 5:e12517
Maheswaran S, Sequist LV, Nagrath S et al (2008) Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 359:366–377
Gandhi J, Zhang J, Xie Y et al (2009) Alterations in genes of the EGFR signaling pathway and their relationship to EGFR tyrosine kinase inhibitor sensitivity in lung cancer cell lines. PLoS One 4:e4576
Wu C, Hao H, Li L et al (2009) Preliminary investigation of the clinical significance of detecting circulating tumor cells enriched from lung cancer patients. J Thorac Oncol 4:30–36
Ray PF, Winston RM, Handyside AH (1996) Reduced allele dropout in single-cell analysis for preimplantation genetic diagnosis of cystic fibrosis. J Assist Reprod Genet 13:104–106
Tewes M, Aktas B, Welt A et al (2009) Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res Treat 115:581–590
Krebs MG, Hou JM, Sloane R et al (2012) Analysis of circulating tumor cells in patients with non-small cell lung cancer using epithelial marker-dependent and -independent approaches. J Thorac Oncol 7:306–315
Zhou X, Li L, Hao H et al (2010) Circulating tumor cells as a prognostic and predictive indicator in advanced lung cancer patients. Oncol Prog 8:484–490
Liu Z, Fusi A, Klopocki E et al (2011) Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J Transl Med 9:70
Yung TKF, Chan KCA, Mok TSK et al (2009) Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 15:2076–2084
Aaltonen KE, Ebbesson A, Wigerup C et al (2011) Laser capture microdissection (LCM) and whole genome amplification (WGA) of DNA from normal breast tissue—optimization for genome wide array analyses. BMC Res Notes 4:69
Pennisi E (2012) The biology of genomes. Single-cell sequencing tackles basic and biomedical questions. Science 336:976–977
Acknowledgments
We thank Zhibin Cheng for help with manuscript preparation. The study was supported by a fund from the Wu Jieping Foundation of China (to L.L).
Conflict of interest
None is declared.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Ran, R., Li, L., Wang, M. et al. Determination of EGFR mutations in single cells microdissected from enriched lung tumor cells in peripheral blood. Anal Bioanal Chem 405, 7377–7382 (2013). https://doi.org/10.1007/s00216-013-7156-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-013-7156-y