Abstract
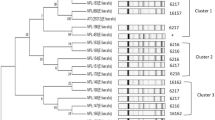
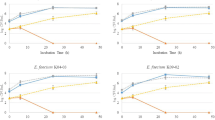
The objective of this study was to characterise lactic acid bacteria (LAB) isolated from faecal samples of healthy Ethiopian infants, with emphasis on bacteriocin production and antibiotic susceptibility. One hundred fifty LAB were obtained from 28 healthy Ethiopian infants. The isolates belonged to Lactobacillus (81/150), Enterococcus (54/150) and Streptococcus (15/150) genera. Lactobacillus species were more abundant in the breast-fed infants while Enterococcus dominated the mixed-fed population. Bacteriocin-producing LAB species were isolated from eight of the infants. Many different bacteriocins were identified, including one new bacteriocin from Streptococcus salivarius, avicin A (class IIa) from Enterococcus avium, one class IIa bacteriocin from Enterococcus faecalis strains, one unknown bacteriocin from E. faecalis and two unknown bacteriocins from Lactobacillus fermentum strains and the two-peptide gassericin T from Lactobacillus gasseri isolate. Susceptibility tests performed for nine antibiotics suggest that some lactobacilli might have acquired resistance to erythromycin (3 %) and tetracycline (4 %) only. The streptococci were generally antibiotic sensitive except for penicillin, to which they showed intermediate resistance. All enterococci were susceptible to ampicillin while 13 % showed penicillin resistance. Only one E. faecalis isolate was vancomycin-resistant. Tetracycline (51 %) and erythromycin (26 %) resistance was prevalent among the enterococci, but multidrug resistance was confined to E. faecalis (47 %) and Enterococcus faecium (33 %). Screening of enterococcal virulence traits revealed that 2 % were β-haemolytic. The structural genes of cytolysin were detected in 28 % of the isolates in five enterococcal species, the majority being E. faecalis and Enterococcus raffinosus. This study shows that bacteriocin production and antibiotic resistance is a common trait of faecal LAB of Ethiopian infants while virulence factors occur at low levels.
Similar content being viewed by others
References
Palmer C, Bik EM, DiGiulio DB et al (2007) Development of the human infant intestinal microbiota. PLoS Biol 5:e177
Penders J, Thijs C, Vink C et al (2006) Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521
Adlerberth I, Wold AE (2009) Establishment of the gut microbiota in Western infants. Acta Paediatr 98:229–238
Orrhage K, Nord CE (1999) Factors controlling the bacterial colonization of the intestine in breastfed infants. Acta Paediatr Suppl 88:47–57
Mitsou EK, Kirtzalidou E, Oikonomou I et al (2008) Fecal microflora of Greek healthy neonates. Anaerobe 14:94–101
Gronlund MM, Lehtonen OP, Eerola E et al (1999) Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr 28:19–25
Ljungh A, Wadstrom T (2006) Lactic acid bacteria as probiotics. Curr Issues Intest Microbiol 7:73–89
Reid G, Jass J, Sebulsky MT (2003) Potential uses of probiotics in clinical practice. Clin Microbiol Rev 16:658–672
Thomas DW, Greer FR (2010) Probiotics and prebiotics in pediatrics. American Academy of Pediatrics 126:1217–1231
Corr SC, Li Y, Riedel CU et al (2007) Bacteriocin production as a mechanism for the antiinfective activity of Lactobacillus salivarius UCC118. Proc Natl Acad Sci U S A 104:7617–7621
EFSA (2012) EFSA panel on additives and products or substances used in animal feed (FEEDAP); guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA Journal 10(6):2740
Salyers AA, Gupta A, Wang Y (2004) Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol 12:412–416
Teuber M, Meile L, Schwarz F (1999) Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Van Leeuwenhoek 76:115–137
Gevers D, Huys G, Swings J (2003) In vitro conjugal transfer of tetracycline resistance from Lactobacillus isolates to other gram-positive bacteria. FEMS Microbiol Lett 225:125–130
Toomey N, Monaghan A, Fanning S et al (2009) Assessment of antimicrobial resistance transfer between lactic acid bacteria and potential foodborne pathogens using in vitro methods and mating in a food matrix. Foodborne Pathog Dis 6:925–933
Jacobsen L, Wilcks A, Hammer K et al (2007) Horizontal transfer of tet(M) and erm(B) resistance plasmids from food strains of Lactobacillus plantarum to Enterococcus faecalis JH2-2 in the gastrointestinal tract of gnotobiotic rats. FEMS Microbiol Ecol 59:158–166
Cataloluk O, Gogebakan B (2004) Presence of drug resistance in intestinal lactobacilli of dairy and human origin in Turkey. FEMS Microbiol Lett 236:7–12
Giraffa G (2002) Enterococci from foods. FEMS Microbiol Rev 26:163–171
Franz CM, Stiles ME, Schleifer KH et al (2003) Enterococci in foods—a conundrum for food safety. Int J Food Microbiol 88:105–122
Murray BE (1990) The life and times of the Enterococcus. Clin Microbiol Rev 3:46–65
Murray BE (1998) Diversity among multidrug-resistant enterococci. Emerg Infect Dis 4:37–47
Moellering RC Jr (1998) Vancomycin-resistant enterococci. Clin Infect Dis 26:1196–1199
Jett BD, Huycke MM, Gilmore MS (1994) Virulence of enterococci. Clin Microbiol Rev 7:462–478
Shankar N, Coburn P, Pillar C et al (2004) Enterococcal cytolysin: activities and association with other virulence traits in a pathogenicity island. Int J Med Microbiol 293:609–618
Abriouel H, Omar NB, Molinos AC et al (2008) Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int J Food Microbiol 123:38–49
Semedo T, Almeida Santos M, Martins P (2003) Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J Clin Microbiol 41:2569–2576
Poeta P, Costa D, Klibi N et al (2006) Phenotypic and genotypic study of gelatinase and beta-haemolysis activities in faecal enterococci of poultry in Portugal. J Vet Med B Infect Dis Vet Public Health 53:203–208
Eaton TJ, Gasson MJ (2001) Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol 67:1628–1635
Mortvedt CI (1990) IF Nes: plasmid-associated bacteriocin production by a Lactobacillus sake strain. J Gen Microbiol 136:1601–1607
Forberg T.(2005) Acid bacteria of different origin, production of antimicrobial substances and distribution of bacteriocin genes. Master Thesis. Norwegian University of Life Sciences, Ås.
Herrera CB. (2006) Faecal bifidobacteria and lactic acid bacteria from breast-fed infants with emphasis on the antimicrobial properties of lactic acid bacteria. Master Thesis. Norwegian University of Life Sciences, Ås.
Booth MC, Bogie CP, Sahl HG et al (1996) Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol Microbiol 21:1175–1184
Jett BD, Jensen HG, Nordquist RE et al (1992) Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect Immun 60:2445–2452
Solheim M, Aakra A, Snipen LG et al (2009) Comparative genomics of Enterococcus faecalis from healthy Norwegian infants. BMC Genomics 10:194
Qin X, Singh KV, Weinstock GM et al (2000) Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun 68:2579–2586
CLSI: Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. CLSI document M100-S22. Clinical and Laboratory Standards Institute, Wayne, PA (2012)
Faye T, Brede DA, Langsrud T et al (2002) An antimicrobial peptide is produced by extracellular processing of a protein from Propionibacterium jensenii. J Bacteriol 184:3649–3656
Bogovic-Matijasic B, Rogelj I, Nes IF et al (1998) Isolation and characterization of two bacteriocins of Lactobacillus acidophilus LF221. Appl Microbiol Biotechnol 49:606–612
Birri DJ, Brede DA, Forberg T et al (2010) Molecular and genetic characterization of a novel bacteriocin locus in Enterococcus avium isolates from infants. Appl Environ Microbiol 76:483–492
Sedgley CM, Clewell DB, Flannagan SE (2009) Plasmid pAMS1-encoded, bacteriocin-related ″Siblicide″ in Enterococcus faecalis. J Bacteriol 191:3183–3188
Kawai Y, Saitoh B, Takahashi O (2000) Primary amino acid and DNA sequences of gassericin T, a lactacin F-family bacteriocin produced by Lactobacillus gasseri SBT2055. Biosci Biotechnol Biochem 64:2201–2208
Birri DJ, Brede DA, Nes IF (2012) Salivaricin D, a novel intrinsically trypsin-resistant lantibiotic from Streptococcus salivarius 5M6c isolated from a healthy infant. Appl Environ Microbiol 78:402–410
Hamilton-Miller JM, Shah S (1998) Vancomycin susceptibility as an aid to the identification of lactobacilli. Lett Appl Microbiol 26:153–154
Edwards CA, Parrett AM (2002) Intestinal flora during the first months of life: new perspectives. Br J Nutr 88(Suppl 1):S11–S18
Dominguez-Bello MG, Costello EK, Contreras M et al (2010) Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107:11971–11975
Martin R, Heilig GH, Zoetendal EG et al (2007) Diversity of the Lactobacillus group in breast milk and vagina of healthy women and potential role in the colonization of the infant gut. J Appl Microbiol 103:2638–2644
Bennet R, Eriksson M, Tafari N et al (1991) Intestinal bacteria of newborn Ethiopian infants in relation to antibiotic treatment and colonisation by potentially pathogenic gram-negative bacteria. Scand J Infect Dis 23:63–69
Stark PL, Lee A (1982) The microbial ecology of the large bowel of breast-fed and formula-fed infants during the first year of life. J Med Microbiol 15:189–203
Yoshioka H, Iseki K, Fujita K (1983) Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics 72:317–321
Ahrne S, Lonnermark E, Wold AE et al (2005) Lactobacilli in the intestinal microbiota of Swedish infants. Microbes Infect 7:1256–1262
Adlerberth I (2008) Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutr Workshop Ser Pediatr Program 62:13–29, discussion 29–33
Hufnagel M, Liese C, Loescher C et al (2007) Enterococcal colonization of infants in a neonatal intensive care unit: associated predictors, risk factors and seasonal patterns. BMC Infect Dis 7:107
Favier CF, Vaughan EE, De Vos WM et al (2002) Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol 68:219–226
O’Shea EF, Gardiner GE, O’Connor PM et al (2009) Characterization of enterocin- and salivaricin-producing lactic acid bacteria from the mammalian gastrointestinal tract. FEMS Microbiol Lett 291:24–34
Garver KI, Muriana PM (1993) Detection, identification and characterization of bacteriocin-producing lactic acid bacteria from retail food products. Int J Food Microbiol 19:241–258
Sedgley CM, Lennan SL, Clewell DB (2004) Prevalence, phenotype and genotype of oral enterococci. Oral Microbiol Immunol 19:95–101
Hallgren A, Claesson C, Saeedi B et al (2009) Molecular detection of aggregation substance, enterococcal surface protein, and cytolysin genes and in vitro adhesion to urinary catheters of Enterococcus faecalis and E. faecium of clinical origin. Int J Med Microbiol 299:323–332
Poeta P, Igrejas G, Costa D et al (2008) Virulence factors and bacteriocins in faecal enterococci of wild boars. J Basic Microbiol 48:385–392
Lopes Mde F, Simoes AP, Tenreiro R (2006) Activity and expression of a virulence factor, gelatinase, in dairy enterococci. Int J Food Microbiol 112:208–214
Coque TM, Patterson JE, Steckelberg JM et al (1995) Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J Infect Dis 171:1223–1229
Cauwerts K, Decostere A, De Graef EM et al (2007) High prevalence of tetracycline resistance in Enterococcus isolates from broilers carrying the erm(B) gene. Avian Pathol 36:395–399
Poeta P, Costa D, Rodrigues J et al (2006) Antimicrobial resistance and the mechanisms implicated in faecal enterococci from healthy humans, poultry and pets in Portugal. Int J Antimicrob Agents 27:131–137
Huys G, D’Haene K, Collard JM et al (2004) Prevalence and molecular characterization of tetracycline resistance in Enterococcus isolates from food. Appl Environ Microbiol 70:1555–1562
Barreto A, Guimaraes B, Radhouani H et al (2009) Detection of antibiotic resistant E. coli and Enterococcus spp. in stool of healthy growing children in Portugal. J Basic Microbiol 49:503–512
Fontana R, Ligozzi M, Pittaluga F et al (1996) Intrinsic penicillin resistance in enterococci. Microb Drug Resist 2:209–213
Hayes JR, English LL, Carr LE et al (2004) Multiple-antibiotic resistance of Enterococcus spp. isolated from commercial poultry production environments. Appl Environ Microbiol 70:6005–6011
EFSA (2012) EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); guidance on the safety assessment of Enterococcus faecium in animal nutrition. EFSA Journal 10(5):2682
Chow JW (2000) Aminoglycoside resistance in enterococci. Clin Infect Dis 31: 586–589
D’Aimmo MR, Modesto M, Biavati B (2007) Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int J Food Microbiol 115:35–42
Temmerman R, Pot B, Huys G et al (2003) Identification and antibiotic susceptibility of bacterial isolates from probiotic products. Int J Food Microbiol 81:1–10
Mandar R, Lijvukene K, Huftt P et al (2001) Antibacterial susceptibility of intestinal lactobacilli of healthy children. Scand J Infect Dis 33:344–349
Belletti N, Gatti M, Bottari B et al (2009) Antibiotic resistance of lactobacilli isolated from two Italian hard cheeses. J Food Prot 72:2162–2169
Klare I, Konstabel C, Werner G (2007) Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J Antimicrob Chemother 59:900–912
Nawaz M, Wang J, Zhou A et al (2011) Characterization and transfer of antibiotic resistance in lactic acid bacteria from fermented food products. Current microbiol 62:1081–1089
Charteris WP, Kelly PM, Morelli L et al (1998) Antibiotic susceptibility of potentially probiotic Lactobacillus species. J Food Prot 61:1636–1643
Delgado S, Florez AB, Mayo B (2005) Antibiotic susceptibility of Lactobacillus and Bifidobacterium species from the human gastrointestinal tract. Curr Microbiol 50:202–207
Weisburg WG, Barns SM, Pelletier DA et al (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Edwards U, Rogall T, Blocker H et al (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853
Stackebrandt E, Charfreitag O (1990) Partial 16S rRNA primary structure of five Actinomyces species: phylogenetic implications and development of an Actinomyces israelii-specific oligonucleotide probe. J Gen Microbiol 136:37–43
Hyink O, Wescombe PA, Upton M et al (2007) Salivaricin A2 and the novel lantibiotic salivaricin B are encoded at adjacent loci on a 190-kilobase transmissible megaplasmid in the oral probiotic strain Streptococcus salivarius K12. Appl Environ Microbiol 73:1107–1113
Xiao H, Chen X, Chen M et al (2004) Bovicin HJ50, a novel lantibiotic produced by Streptococcus bovis HJ50. Microbiology 150:103–108
Shankar N, Baghdayan AS, Gilmore MS (2002) Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746–750
Martinez-Bueno M, Maqueda M, Galvez A et al (1994) Determination of the gene sequence and the molecular structure of the enterococcal peptide antibiotic AS-48. J Bacteriol 176:6334–6339
Tomita H, Fujimoto S, Tanimoto K et al (1996) Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pYI17. J Bacteriol 178:3585–3593
Balla E, Dicks LM, Du Toit M et al (2000) Characterization and cloning of the genes encoding enterocin 1071A and enterocin 1071B, two antimicrobial peptides produced by Enterococcus faecalis BFE 1071. Appl Environ Microbiol 66:1298–1304
Acknowledgements
Grants from Norwegian Research Council have supported this study. We thank Zhian Salehian for technical assistance. We acknowledge parents for allowing us to collect samples from their infants.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Birri, D.J., Brede, D.A., Tessema, G.T. et al. Bacteriocin Production, Antibiotic Susceptibility and Prevalence of Haemolytic and Gelatinase Activity in Faecal Lactic Acid Bacteria Isolated from Healthy Ethiopian Infants. Microb Ecol 65, 504–516 (2013). https://doi.org/10.1007/s00248-012-0134-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-012-0134-7