Abstract
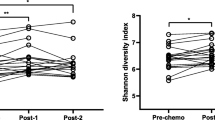
Gastrointestinal disturbances are a side-effect frequently associated with haematological malignancies due to the intensive cytotoxic treatment given in connection with bone marrow transplantation (BMT). However, intestinal microbiota changes during chemotherapy remain poorly described, probably due to the use of culture-based and low-resolution molecular methods in previous studies. The objective of our study was to apply a next generation DNA sequencing technology to analyse chemotherapy-induced changes in faecal microbiota. We included eight patients with non-Hodgkin’s lymphoma undergoing one course of BMT conditioning chemotherapy. We collected a prechemotherapy faecal sample, the day before chemotherapy was initiated, and a postchemotherapy sample, collected 1 week after the initiation of chemotherapy. Total DNA was extracted from faecal samples, denaturing high-performance liquid chromatography based on amplification of the V6 to V8 region of the 16S ribosomal RNA (rRNA) gene, and 454-pyrosequencing of the 16 S rRNA gene, using PCR primers targeting the V5 and V6 hypervariable 16S rRNA gene regions were performed. Raw sequence data were screened, trimmed, and filtered using the QIIME pipeline. We observed a steep reduction in alpha diversity and significant differences in the composition of the intestinal microbiota in response to chemotherapy. Chemotherapy was associated with a drastic drop in Faecalibacterium and accompanied by an increase of Escherichia. The chemotherapy-induced shift in the intestinal microbiota could induce severe side effects in immunocompromised cancer patients. Our study is a first step in identifying patients at risk for gastrointestinal disturbances and to promote strategies to prevent this drastic shift in intestinal microbiota.





Similar content being viewed by others
References
Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR et al (2008) Evolution of Mammals and Their Gut Microbes. Science 320:1647–1651
Dethlefsen L, McFall-Ngai M, Relman DA (2007) An Ecological and Evolutionary Perspective on Human–Microbe Mutualism and Disease. Nature 449:811–818
Ley RE, Peterson DA, Gordon JI (2006) Ecological and Evolutionary Forces Shaping Microbial Diversity in the Human Intestine. Cell 124:837–848
Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R et al (2007) The Human Microbiome Project. Nature 449:804–810
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L et al (2005) Diversity of the Human Intestinal Microbial Flora. Science 308:1635–1638
Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS et al (2009) Characterizing a Model Human Gut Microbiota Composed of Members of Its Two Dominant Bacterial Phyla. Proc Natl Acad Sci U S A 106:5859–5864
Guarner F, Malagelada JR (2003) Gut Flora in Health and Disease. Lancet 361:512–519
Stringer AM, Gibson RJ, Logan RM, Bowen JM, Yeoh AS et al (2008) Fecal Microflora and beta-Glucuronidase Expression are Altered in an Irinotecan-Induced Diarrhea Model in Rats. Cancer Biol Ther 7:1919–1925
Stringer AM, Gibson RJ, Bowen JM, Logan RM, Ashton K et al (2009) Irinotecan-Induced Mucositis Manifesting as Diarrhoea Corresponds with an Amended Intestinal Flora and Mucin Profile. Int J Exp Path 90:489–499
van Vliet MJ, Tissing WJ, Dun CA, Meessen NE, Kamps WA et al (2009) Chemotherapy Treatment in Pediatric Patients with Acute Myeloid Leukemia Receiving Antimicrobial Prophylaxis Leads to a Relative Increase of Colonization with Potentially Pathogenic Bacteria in the Gut. Clin Infect Dis 49(2):262–270
Zwielehner J, Lassl C, Hippe B, Pointner A, Switzeny OJ et al (2011) Changes in Human Fecal Microbiota due to Chemotherapy Analyzed by TaqMan-PCR, 454 Sequencing and PCR-DGGE Fingerprinting. Plos One 6:e28654
Goldenberg O, Herrmann S, Marjoram G, Noyer-Weidner M, Hong G et al (2007) Molecular Monitoring of the Intestinal Flora by Denaturing High Performance Liquid Chromatography. J Microbiol Methods 6894–105
Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ et al (2006) Metagenomic Analysis of the Human Distal Gut Microbiome. Science 312:1355–1359
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER et al (2006) An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature 444:1027–1031
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A et al (2009) A Core Gut Microbiome in Obese and Lean Twins. Nature 457:480–484
Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R et al (2009) The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci Transl Med 1:6–14
Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ et al (2012) Inflammasome-Mediated Dysbiosis Regulates Progression of NAFLD and Obesity. Nature 482:179–185
Zoetendal EG, Rajilic-Stojanovic M, de Vos WM (2008) High-Throughput Diversity and Functionality Analysis of the Gastrointestinal Tract Microbiota. Gut 57:1605–1615
Dethlefsen L, Huse S, Sogin ML, Relman DA (2008) The Pervasive Effects of an Antibiotic on the Human Gut Microbiota, As Revealed by Deep 16S rRNA Sequencing. Plos Biol 6:e280
Grice EA, Kong HH, Conlan S, Deming CB, Davis J et al (2009) Topographical and Temporal Diversity of the Human Skin Microbiome. Science 324:1190–1192
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS et al (2010) A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature 464:59–65
Arnold RJ, Gabrail N, Rat M, Kim R, Sung JC et al (2005) Clinical Implications of Chemotherapy Induced Diarrhea in Patients with Cancer. J Support Oncol 3:227–232
van Vliet MJ, Harmsen HJ, de Bont ES, Tissing WJ (2010) The Role of Intestinal Microbiota in the Development and Severity of Chemotherapy-Induced Mucositis. Plos Pathol 27:e1000879
Sonis ST (2004) The Pathobiology of Mucositis. Nat Rev Cancer 4:277–284
Von Bültzingslöwen I, Adlerberth I, Wold AE, Dahlén G, Jontell M (2003) Oral and Intestinal Microflora in 5-Fluorouracil Treated Rats, Translocation to Cervical and Mesenteric Lymph Nodes and Effects of Probiotic Bacteria. Oral Microbiol Immunol 18:278–284
Dethlefsen L, Relman DA (2011) Incomplete Recovery and Individualized Responses of the Human Distal Gut Microbiota to Repeated Antibiotic Perturbation. Proc Natl Acad Sci U S A 108:4554–4561
Schneider SM, Le Gall P, Girard-Pipau F, Piche T, Pompei A et al (2000) Total Artificial Nutrition Is Associated With Major Changes in the Fecal Flora. Eur J Nutr 39:248–255
Fishman JA (1998) Prevention of Infection due to Pneumocystis carinii. Antimicrob Agents Chemother 42(5):995–1004
de La Cochetiere MF, Durand T, Lepage P, Bourreille A, Galmiche JP et al (2005) Resilience of the Dominant Human Fecal Microbiota upon Short-Course Antibiotic Challenge. J Clin Microbiol 43:5588–5592
Caillaux G, de La Cochetière MF, Carton T, Le Vacon F, Rozé JC et al (2010) Application of Denaturing High-Performance Liquid Chromatography for Intestinal Microbiota Analysis of Newborns. J Perinat Med 38:339–341
Rougé C, Goldenberg O, Ferraris L, Berger B, Rochat F et al (2010) Investigation of the Intestinal Microbiota in Preterm Infants Using Different Methods. Anaerobe 16:362–370
Andersson AF, Lindberg M, Jakobsson H, Bäckhed F, Nyrén P et al (2008) Comparative Analysis of Human Gut Microbiota by Barcoded Pyrosequencing. Plos One 30:e2836
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD et al (2010) QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat Methods 7:335–336
Wang QG, Garrity M, Tiedje JM, Cole JR (2007) Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol 73(16):5261–5267
Lozupone C, Knight R (2005) Unifrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol 71(12):8228–8235
Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D et al (2010) Metagenomic Analyses Reveal Antibiotic-Induced Temporal and Spatial Changes in Intestinal Microbiota with Associated Alterations in Immune Cell Homeostasis. Mucosal Immunol 3:148–158
Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG et al (2008) Faecalibacterium prausnitzii Is an Anti-Inflammatory Commensal Bacterium Identified by Gut Microbiota Analysis of Crohn Disease Patients. Proc Natl Acad Sci U S A 105:16731–16736
Montassier E, Batard E, Gastinne T, Potel G, de La Cochetière MF (2013) Recent Changes in Bacteremia in Patients with Cancer: A Systematic Review of Epidemiology and Antibiotic Resistance. Eur J Clin Microbiol Infect Dis
Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I et al (2009) Low Counts of Faecalibacterium prausnitzii in Colitis Microbiota. Inflamm Bowel Dis 15:1183–1189
Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML et al (2010) Bifidobacterium animalis subsp. lactis Fermented Milk Product Reduces Inflammation by Altering a Niche for Colitogenic Microbes. Proc Natl Acad Sci U S A 107(42):18132–18137
Borchers AT, Selmi C, Meyers FJ, Keen CL, Gershwin ME (2009) Probiotics and Immunity. J Gastroenterol 44:26–46
Bodet CA 3rd, Jorgensen JH, Drutz DJ (1985) Antibacterial Activities of Antineoplastic Agents. Antimicrob Agents Chemother 28(3):437–439
Hamilton-Miller JM (1984) Antimicrobial Activity of 21 Anti-neoplastic Agents. Br J Cancer 49(3):367–369
Durbán A, Abellán JJ, Jiménez-Hernández N, Ponce M, Ponce J et al (2011) Assessing Gut Microbial Diversity from Feces and Rectal Mucosa. Microb Ecol 61(1):123–133
Pilloni G, Granitsiotis MS, Engel M, Lueders T (2012) Testing the Limits of 454 Pyrotag Sequencing: Reproducibility, Quantitative Assessment and Comparison to T-RFLP Fingerprinting of Aquifer Microbes. PLoS One 7(7):e40467
Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP (2013) Stool Microbiome and Metabolome Differences between Colorectal Cancer Patients and Healthy Adults. PLoS One 8(8):e70803
Wu N, Yang X, Zhang R, Li J, Xiao X et al (2013) Dysbiosis Signature of Fecal Microbiota in Colorectal Cancer Patients. Microb Ecol 66(2):462–470
Chen HM, Yu YN, Wang JL, Lin YW, Kong X et al (2013) Decreased Dietary Fiber Intake and Structural Alteration of Gut Microbiota in Patients with Advanced Colorectal Adenoma. Am J Clin Nutr 97(5):1044–1052
Knox C, Law V, Jewison T, Liu P, Ly S et al (2011) Drugbank 3.0: A Comprehensive Resource for ‘Omics’ Research on Drugs. Nucleic Acids Res 39:D1035–D1041
Acknowledgments
This work was funded by the Nantes University Hospital Grant BRD/10/04-Q and by the Mérieux Research Grants.
Accession Number
The V5–V6 16S rDNA bacterial sequences analysed in this paper have been deposited in the GenBank Short Read Archive (Accession number: SRA116522).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
SI Figure 1
Denaturing high-performance liquid chromatography (dHPLC) fingerprint analysis of the V6 to V8 region of the 16S rRNA gene. Example of dHPLC fingerprints obtained in one patient, S1: prechemotherapy sample, S2: postchemotherapy sample. The dominant microbiota was converted into a profile with peaks, each peak corresponds to an amplicon. (JPEG 20 kb)
SI Figure 2
Most abundant phyla found in the prechemotherapy (S1) and postchemotherapy (S2) faecal samples of eight patients (p1 to p8) undergoing bone marrow transplantation conditioning chemotherapy. (JPEG 102 kb)
SI Figure 3
Most abundant genera found in the prechemotherapy (S1) and postchemotherapy (S2) faecal samples of 8 patients (p1 to p8) undergoing bone marrow transplantation conditioning chemotherapy. (JPEG 165 kb)
SI Table 1
OTUs that were significantly different in abundance before and after chemotherapy. (DOC 88 kb)
Rights and permissions
About this article
Cite this article
Montassier, E., Batard, E., Massart, S. et al. 16S rRNA Gene Pyrosequencing Reveals Shift in Patient Faecal Microbiota During High-Dose Chemotherapy as Conditioning Regimen for Bone Marrow Transplantation. Microb Ecol 67, 690–699 (2014). https://doi.org/10.1007/s00248-013-0355-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-013-0355-4