Abstract
Mutations in PROP1 are a common genetic cause of multiple pituitary hormone deficiency (MPHD). We used a comparative genomics approach to predict the transcriptional regulatory domains of Prop1 and tested them in cell culture and mice. A BAC transgene containing Prop1 completely rescues the Prop1 mutant phenotype, demonstrating that the regulatory elements necessary for proper PROP1 transcription are contained within the BAC. We generated DNA sequences from the PROP1 genes in lemur, pig, and five different primate species. Comparison of these with available human and mouse PROP1 sequences identified three putative regulatory sequences that are highly conserved. These are located in the PROP1 promoter proximal region, within the first intron of PROP1, and downstream of PROP1. Each of the conserved elements elicited orientation-specific enhancer activity in the context of the Drosophila alcohol dehydrogenase minimal promoter in both heterologous and pituitary-derived cells lines. The intronic element is sufficient to confer dorsal expansion of the pituitary expression domain of a transgene, suggesting that this element is important for the normal spatial expression of endogenous Prop1 during pituitary development. This study illustrates the usefulness of a comparative genomics approach in the identification of regulatory elements that may be the site of mutations responsible for some cases of MPHD.
Similar content being viewed by others
Introduction
All vertebrates have pituitary glands composed of specialized hormone-producing cells (Matsumoto and Ishii 1987). The individual hormones are evolutionarily conserved, although their function varies across the classes of Animalia. This conservation suggests that genetic regulation of pituitary function may be conserved.
In humans, growth insufficiency resulting from pituitary hormone deficiency is not infrequent, occurring in approximately 1 in 4000 live births (Procter et al. 1998; Vimpani et al. 1977). Growth hormone (GH) insufficiency is the most common type of dwarfism and usually results from mutations in the GH gene cluster (Braga et al. 1986; Mullis et al. 1990). Multiple pituitary hormone deficiencies (MPHD) result from mutations in transcription factors important for the normal development and function of the pituitary gland, including POU1FI (PIT1), Prophet of PIT1 (PROP1), HESX1, LHX3, LHX4, and SOX3 (Bhangoo et al. 2006; Cogan et al. 1998; Laumonnier et al. 2002; Machinis et al. 2001; Mendonca et al. 1999; Netchine et al. 2000; Pfäffle et al. 1992; Radovick et al. 1992; Tajima et al. 2003; Wu et al. 1998). The first transcription factor to be linked to MPHD was PIT1 (Tatsumi et al. 1992). Patients with mutations in PIT1 generally have deficiencies in GH, prolactin (PRL), and thyroid-stimulating hormone (TSH) as well as profound pituitary hypoplasia (Cohen et al. 1996). Mutations in PROP1 are a common genetic cause of familial MPHD. Patients with PROP1 mutations exhibit progressive hormone loss with varying age of onset and severity (Bottner et al. 2004; Fluck et al. 1998). Most common are deficiencies in PRL, GH, and TSH as well as follicle-stimulating hormone (FSH) and luteinizing hormone (LH) (Agarwal et al. 2000; Deladoey et al. 1999). Progressive adrenocorticotrophic hormone (ACTH) loss presents as late as the third decade of life (Bottner et al. 2004). Some MPHD cases, however, cannot be traced to mutations in the protein-coding or intron-exon splice sites in the DNA sequence of known genes important for pituitary development.
The mechanism of PROP1 action has been studied extensively in two mouse models, the Ames dwarf (Prop1df) and the Prop1null (Nasonkin et al. 2004; Ward et al. 2005). In mice, Prop1 is expressed throughout the developing Rathke’s pouch in a dorsal to ventral expression gradient from about embryonic day 10 (e10) until about e16 (Sornson et al. 1996). PROP1 transcripts are present in the adult pituitaries of human and pig, although the levels were not quantified relative to the embryonic pituitary (Nakamura et al. 1999a, 1999b; Skelly et al. 2000; Sloop et al. 2000; Usui et al. 2000). A number of Prop1 downstream targets have been identified, including Pit1, Hesx1, Tle3, and Notch2 (Brinkmeier et al. 2003; Gage et al. 1996a; Raetzman et al. 2004).
It is of great interest to identify the transcriptional regulators of Prop1 because it is pituitary-specific, unlike many of the other key regulators of pituitary development: Pitx1, Pitx2, Lhx3, Lhx4, and Hesx1. Each of these genes is expressed prior to Prop1 and is a candidate for transcriptional regulation of Prop1. Prop1 expression is activated in Pitx1 and Pitx2 single mutants, but this may be due to the ability of Pitx1 and Pitx2 to compensate for each other (Suh et al. 2002; Szeto et al. 1999). Prop1 expression also appears to be initiated normally in Hesx1- and Lhx4-deficient mice (Dasen et al. 2001; Raetzman et al. 2002). Thus, the spatial and temporal regulation of Prop1 expression is not fully explained by these genes, suggesting that additional factors may be involved.
The regulation of gene expression involves the cooperation of a variety of transcription factors in a tissue-, temporal-, and/or spatial-specific fashion that interact with cis-acting regulatory elements in DNA sequences (Kleinjan and van Heyningen 2005). The identification of these elements can be difficult because they may be located at a considerable distance from the gene or even within the introns of neighboring genes (Bagheri-Fam et al. 2001; Lang et al. 2003; Lettice et al. 2003). The identification of these elements is facilitated by comparative genomics in the form of cross-species DNA comparisons. The alignment of the DNA sequences of orthologous genes from different species, both closely and distantly related, can reveal potential conserved regulatory elements that can then be analyzed in vivo (Boffelli et al. 2004; Nobrega and Pennacchio 2004; Pennacchio and Rubin 2001). Transcription factors involved in development, like Prop1, are often conserved among vertebrates, and their regulatory sequences are also likely to be conserved (Plessy et al. 2005).
In this study we report the sequence of the PROP1 gene from several mammals and utilize cross-species PROP1 protein sequence comparison to verify the conservation of the functional domains of the protein and use genomic sequence comparison to identify putative transcriptional regulatory elements in the noncoding regions of the Prop1 gene. This analysis revealed the presence of three conserved noncoding elements within and near the gene. Each of them exhibited orientation-dependent enhancer activity in tissue culture, and an element in intron 1 conferred tissue-specific and unique spatial expression in transgenic mice. These studies establish a functional role for the intronic element in Prop1 gene regulation. Finally, using bacterial artificial chromosome (BAC) transgene rescue of the Prop1 mutant phenotype, we demonstrate that all of the sequences necessary for functional expression of Prop1 are located within the BAC.
Materials and methods
Protein and DNA sequence
A BAC clone of approximately 200 kb and containing the pig (Sus scrofa) Prop1 gene was identified, and DNA was isolated from it. A shotgun sequencing library was prepared from that BAC by using SeqWright (Houston, TX), and the resulting subclones were sequenced at the University of Michigan Sequencing Core using Applied Biosystems (Foster City, CA) sequencers (Model 3700) and BigDye V1.1 terminator chemistry, according to standard manufacturer’s protocols. Sequence assembly of the shotgun sequence data was performed using phred and phrap (Ewing and Green 1998), and consed (Gordon et al. 1998), resulting in 6X draft sequence coverage. Limited finishing was performed based on the “autofinish” option in consed to close some gaps, especially those in the vicinity of the Prop1 gene. A total of ten draft contigs of 1 kb or greater were obtained in the final assembly, accounting for 196 kb, or an estimated 98% of the original BAC. The sequences have a phred Q-score of 20 or higher, with the majority being Q40 or better. The BAC sequences were submitted to GenBank (NCBI; http://www.ncbi.nlm.nih.gov accession number EF590118). One contig of approximately 26.3 kb contained the Sus scrofa Prop1 gene, including 13.8 kb of 5′ flanking sequence, 3.7 kb encoding the Prop1 gene, and 8.7 kb of 3′ flanking sequence. A set of lemur BACs containing the Prop1 gene were isolated from a library derived from a cell line of the ring-tailed lemur (Lemur catta; AG07100C, Coriell Cell Repositories, Camden, NJ) using a human PROP1 exon 3 probe. One lemur BAC, LBNL-2 102B17, was sequenced from ends of 3 kb subclones to approximately tenfold coverage using BigDye terminators (Applied Biosystems) and assembled into ordered and oriented contigs with the Phred-Phrap-Consed suite (Ewing and Green 1998; Gordon et al. 1998). The assembled BAC sequence was submitted to GenBank with accession number AC162436. Prop1 gene sequence from gorilla (Gorilla gorilla), Black and Red Howler (Alouatta belzebul), Brown Capuchin (Cebus apella), and Gelada Baboon (Theropithecus gelada) was amplified from genomic DNA (gift from Dr Deborah Gumucio, University of Michigan, Ann Arbor, MI) by using the forward primer (5′-CCTGCTCCCAGGAGGGGATT-3′) that corresponded to the human PROP1 5′ flanking sequence that was highly conserved between mouse and human, and as the reverse primer (5′-AGGCTGGGGATCACCTTGGTG-3′) that corresponded to the 3′ UTR of the human PROP1 gene. cDNA sequence was determined by using the high conservation between primate exon splice acceptor/donor sites to assemble the cDNA sequence from the gene sequence. The protein sequence was then translated from the cDNA as described above. The Prop1 gene sequences were deposited in GenBank with accession numbers DQ177426 for gorilla, DQ177425 for capuchin monkey, DQ177427 for howler monkey, and DQ177424 for baboon.
The 20-kb human PROP1 was obtained from the human chromosome 5 contig sequences from GenBank accession numbers NT086684 and NT023133. The 20-kb mouse (Mus musculus) Prop1 sequence was obtained from the mouse chromosome 11 contig sequences from GenBank accession number NT096135. The Prop1 genomic sequence for the chimpanzee (Pan troglodytes) was obtained via the Berkley Genome Pipeline from the genome VISTA analysis program [http://www.genome.lbl.gov/vista/index.shtml (Couronne et al. 2003)]. The cDNA and protein sequences were determined as described above. The Prop1 genomic sequence for the rat and partial genomic sequence for the fugu (Fugu rubripes) and zebrafish (Danio rerio) were obtained by searching the UCSC genome browser [http://www.genome.ucsc.edu (Kent 2002)] for the closest matches to PROP1. The partial protein sequences for the fugu and zebrafish were determined by the translation of the partial gene sequence in all three frames to identify the PROP1 homedomain and transactivation domain sequence. The rat (Rattus norvegicus) cDNA sequence was obtained from GenBank (accession number NM153627) and translated as described above. Prop1 genomic sequence was obtained for the dog (Canis familiaris; AF126157) and sheep (Ovis aries; AY533708) and PROP1 protein sequence for human (NP006252), pig (NP001001263), cow (Bos taurus; NP777103), sheep (NP001009767), mouse (P97458), dog (NP001018643), and partial protein sequence for the chicken (Gallus gallus; AB037110) was obtained from the NCBI website.
Sequence analysis
ClustalW alignment for protein and DNA sequences were done with the LASERGENE Navigator Meg align sequence alignment program (DNASTAR Inc., Madison, WI). Mouse, rat, human, and chimp chromosome comparisons were done using the University of California, Santa Cruz (UCSC) genome browser [http://www.genome.ucsc.edu (Kent et al. 2002)]. The pig Prop1 BAC contigs were compared to the mouse genome using the genome VISTA program [http://www.genome.lbl.gov/vista/index.shtml (Couronne et al. 2003)]. The 20-kb PROP1 genomic sequences for the human, lemur, pig, and mouse were analyzed with the mVISTA [http://www.genome.lbl.gov/vista/index.shtml (Bray et al. 2003)] comparative genomics program to determine the identity of the conserved noncoding elements.
Plasmid construction
The CE-A/LacZ plasmid was constructed for the targeted knockout of the Prop1 gene (Nasonkin et al. 2004). The CE-B + CE-A/LacZ plasmid was constructed by digesting the CE-A/LacZ plasmid with XhoI and subcloning the region containing 3 kb of the Prop1 5′ flanking region, the LacZ coding region, and mouse protamine 1 splice and polyadenylation regions into the XhoI site of pBluescript SK+ (Stratagene, La Jolla, CA). A 9.5-kb Prop1 genomic clone (Nasonkin et al. 2004), which was generated from a P1 clone containing the entire Prop1 gene, was used as a template to amplify the CE-B region with a series of primers that engineered flanking loxP sites (5′-ATAACTTCGTATAGCATACATTATACGAAGTTAT-3′). The floxed CE-B fragment was subcloned into pGEM-T Easy (Promega, Madison, WI). This CE-B loxP construct was digested with NotI/XbaI and ligated into the NotI/XbaI sites of the pBluescript SK+ vector that contained the 3-kb Prop1 5′ flanking sequence with the LacZpA reporter.
The αGsu-Prop1ΔCE-B plasmid was made by creating a chimera of the Prop1 intron 1 in which the CE-B region was deleted. First, the intron sequence for the 5′ flank of CE-B was amplified from the 9.5-kb Prop1 genomic clone (Nasonkin et al. 2004) by using primers (5′-GGTTTGGGTGGCTAGCCATGGAA-3′ and 5′-TTCCCAAGCACCTCCTTCATATCCCACCCCCCAACTAAGCACCC-3′) that allowed this fragment to be annealed to the CE-B 3′ flanking sequence that was also amplified from the 9.5-kb Prop1 plasmid with the primers 5′-CCTCCTATAAGCCTCAGAGCT-3′ and 5′-GGGTGCTTAGTTGGGGGGTGGGATATGAAGGAGGTGCTTGGG-3′. These two PCR products were engineered with overlapping tails that could be annealed together to create a chimeric Prop1 intron1 with the CE-B region deleted. This chimera was amplified with the primers 5′-GGTTTGGGTGGCTAGCCATGGAA-3′ and 5′-CCTCCTATAAGCCTCAGAGCT-3′, digested with NcoI and SacI, and subcloned into the αGsu-Prop1 plasmid (Cushman et al. 2001) to create the desired transgenic construct. This αGsu-Prop1ΔCE-B was subcloned into pBluescript SK+ by an EcoR1 partial digest. The CE-B + αGsu-Prop1ΔCE-B plasmid was made by amplifying the CE-B loxP region from the CE-B + CE-A/LacZ plasmid with the primers 5′-GGTATCGATTACCCTAGAGGGCAGTGCAGTGCCTG-3′ and 5′-GGAATCGATATCTCTTTGCTGTCTATCAATGACGT-3′ that engineered ClaI sites at the ends of the PCR product to allow the subcloning of this CE-B loxP region into the ClaI site of αGsu-Prop1ΔCE-B.
The 584-bp CE-A region was amplified from the 9.5-kb Prop1 genomic clone with primers to engineer HindIII sites at the ends the sequence (5′-GTCTGGAAGCTTGCTGGTGAGGCTG-3′ and 5′-GGAAGCTTGTCTTGGAGAAGAGACCTCCTCCTGG-3′) and subcloned in both the forward and reverse orientations into the HindIII site of the pDeltaODLO 02 plasmid (Iniguez-Lluhi et al. 1997) obtained from Dr. Jorge Iniguez-Lluhi (University of Michigan, Ann Arbor, MI) that contained the Drosophila alcohol dehydrogenase (ADH) minimal promoter and the firefly luciferase reporter gene. The 508-bp CE-B region was PCR amplified from the 9.5-kb Prop1 plasmid with primers that engineered EcoRI sites at the ends (5′-CGGAAGAATTCTGGTTGCCCAAGGTCC-3′ and 5′-GCCACTCGCAGAATTCATTTC-3′) and subcloned into pBluescript (Stratagene) at the EcoRI site. The CE-B was then digested with SmaI/KpnI and subcloned onto a version of pGL3basic (Promega) that contained the TK minimal promoter inserted into the BglI/HindIII sites. The CE-B region was released from this plasmid by digestion with XhoI and subcloned into the XhoI site of pDeltaODLO 02 in both the reverse and forward orientations. The 1196-bp CE-C region was amplified from the 9.5-kb plasmid (5′-GGAGTACTGGGACCCTTAAGGCCCTTGGGCTGCAGG-3′ and 5′-GGAGTACTGGAGTCTGAGACAGGAAGACTGAGAG-3′), cloned in to the pGEM-T Easy vector (Promega), digested with NotI, and subcloned into the NotI site of pDeltaODLO 02 in the forward and reverse orientations.
Cell culture
Monkey fibroblast CV-1 cells (American Type Culture Collection, Manassas, VA), mouse pituitary gonadotrope αT3-1 cells (Dr. Pamela Mellon, University of San Diego, La Jolla, CA), rat anterior pituitary GH3 cells, and mouse pituitary corticotrope AtT-20 cells were maintained at 37°C/5% CO2 in Dulbecco’s Modified Eagle Medium (Invitrogen, Carlsbad, CA) supplemented with 10% heat inactivated fetal bovine serum (Hyclone, Logan, UT) and 100 units/ml penicillin-streptomycin (Invitrogen). The GH3 and AtT-20 cell lines were obtained from Dr. Audrey Seasholtz (University of Michigan, Ann Arbor, MI).
Transient transfection and Dual-Luciferase® Reporter Assay
Cells were plated onto 24-well plastic plates (Fisher Scientific, Fair Lawn, NJ) at a density of 0.4 × 105 cells/well for CV-1, 0.7 × 105 cells/well for αT3-1, 1.0 × 105 cells/well for GH3, and 1.2 × 105 cells/well for AtT-20 cells, such that cells were 40%–60% confluent the next day. DNA cocktails totaling 0.3 μg/well [0.08 μg enhancer construct, 0.218 μg pBluescript SK+, 0.002 μg (cytomegalovirus) CMV-Renilla luciferase (Promega) internal control in 400 μl serum-free DMEM] were transfected into cultured cells using Fugene 6 (Roche, Indianapolis, IN) at a 12:5 ratio according to the manufacturer’s protocol. The pDeltaODLO 02 plasmid that contains the ADH minimal promoter with the firefly luciferase reporter gene was used as a negative control and determined as basal level. Forty-eight hours after transfection, Dual-Luciferase Reporter Assay (Promega) was performed according to the manufacturer’s protocol and measured using the Lmax Micro plate Illuminometer (Molecular Devices, Sunnyvale, CA) with the SOFTmax Pro software (Molecular Devices). The results were normalized to the CMV-Renilla luciferase internal control. All assays were done in triplicate and the results were repeated a total of three times. Results were averaged and expressed as percent activity over basal.
Maintenance, generation, and genotyping of transgenic mice
To generate the BAC transgenic mice, BAC RP23-250I22 (supplied by Pieter J. de Jong, Childrens Hospital Oakland Research Institute) was purified over a Nucleobond AX column (BD (Biosciences) and injected into pronuclei of fertilized eggs generated from a cross between DF/B-Prop1 df/+ males of mixed genetic background (Buckwalter et al. 1991) and (SJL/J × C57BL/6J) F1 females. BAC transgenic; Prop1 df/+ mice were crossed to N6-B6-Prop1 +/− mice (Nasonkin et al. 2004) to generate BAC transgenic; Prop1 df/- offspring. These mice and all littermates were weighed, photographed, and genotyped at weaning.
The presence of the BAC was assessed by PCR using primers designed to amplify products that span the junction between the BAC backbone and the mouse genomic DNA (Sp6 end 5′-CATATTTTCCCCATCCACCACCAT-3′ and 5′-TTCCCGCAAGAGCAAACACAAC-3′; T7 end 5′-CCGGAAGGAGCTGACTGGGTTGA-3′ and 5′-TGGGCATTGAGCTTTCTGGGTTTT-3′). Previously described primers were used to genotype the Prop1 df allele (Cushman et al. 2001) and Prop1 null allele (Nasonkin et al. 2004).
The αGSU Prop1 transgenic mouse lines TgN(Cga-Prop1)D4Sac and TgN(Cga-Prop1)D6Sac (Cushman et al. 2001; Vesper et al. 2006) have been maintained in the mouse facility at the University of Michigan. Newborns were obtained by mating transgenic males of the D4 or D6 αGsu-Prop1 transgenic lines with C57BL/6J females (The Jackson Laboratory, Bar Harbor, ME). Genomic DNA was prepared from tail biopsies of all progeny born and then screened for the αGsu-Prop1 transgene as previously described (Cushman et al. 2001).
To create transient transgenic mice with various Prop1 plasmids, inserts were released from the plasmid vector sequences and purified for microinjection. The 7.7-kb αGsu-Prop1ΔCE-B fragment was generated by digestion of the αGsu-Prop1ΔCE-B plasmid with NotI/ClaI. The 8.0-kb CE-B + αGsu-Prop1ΔCE-B fragment was generated by the digestion of the CE-B + αGsu-Prop1ΔCE-B plasmid with NotI/ApaI. Both inserts were isolated by agarose gel electrophoresis and purified with the Nucleospin Extract Kit (Clontech, Mountain View, CA). Microinjection and transplantation were performed as previously reported (Cushman et al. 2001). Genomic DNA was prepared from tail biopsies of all progeny born and then screened for the transgene using the same genotyping strategy as for the αGsu-Prop1 (Cushman et al., 2001).
The 7-kb CE-A/LacZ fragment was generated by digestion of the CE-A/LacZ plasmid with XhoI. The 8.5-kb CE-B + CE-A/LacZ plasmid was generated by digestion of the CE-B + CE-A/LacZ plasmid with NotI/ScaI. Microinjection and transplantation were performed as described above. To detect both transgenes, a 250-bp product was amplified from the genomic DNA using the Prop1-specific primer (5′-GTGAGAAAACAGGTATCTAGCT-3′) and the LacZ-specific primer (5′-CCACTTTGCGTTTCTTGG-3′). Reactions were performed for 33 cycles of PCR conditions: 93°C for 3 min × 1, (94°C for 30 sec, 55°C for 45 sec, 72°C for 20 sec), 72°C for 5 min.
All mice were housed in a 12-h light, 12-h dark cycle with unlimited access to tap water and Purina 5008 or 5020 chow. All procedures using mice were approved by the University of Michigan Committee on Use and Care of Animals, and all experiments were conducted in accordance with the principles and procedures outlined in the NIH Guidelines for the Care and Use of Experimental Animals.
Analysis of transgenic animals
Embryos were harvested on e12.5 from surrogate mothers carrying CE-B + CE-A/LacZ transient transgenics and quick frozen on dry ice. Cryosections of 12–15 μm were prepared on slides and fixed in 0.5% glutaraldehyde, 1.25 mM EGTA, 2 mM MgCl2, and PBS (pH 7.2) for 5 min at room temperature, washed three times in 0.02% NP-40 (Amersham Pharmacia Biotech, Inc., Piscataway, NJ), 100 mM sodium phosphate, 2 mM MgCl2 wash buffer, and stained for β-galactosidase activity overnight at 37°C in a solution of 1 mg/ml X-gal (Roche), 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, 2 mM MgCl2, and 0.02% NP-40 in PBS.
P1 heads from αGsu-Prop1, αGsu-Prop1ΔCE-B, CE-B+αGsu-Prop1ΔCE-B, and nontransgenic controls; e12.5 embryos from CE-A/LacZ, CE-B+CE-A/LacZ, and controls; or e12.5/e14.5 embryos from wild-type animals were harvested and fixed for 2-24 h in 4% paraformaldehyde in PBS (pH 7.2) followed by PBS wash, dehydration in a graded series of ethanol, and paraffin embedding. Six-micrometer sections were prepared on slides and washed in 0.3% Triton X-100 (Sigma, St. Louis, MO) in PBS (pH 7.2) for 15 min at room temperature, permeablized by proteinase K digestion (0.8 μg/ml in 100 mM Tris-HCl, 50 mM EDTA pH 8.0) for 15 min at 37°C, followed by a 5-min fixation in 4% paraformaldehyde in PBS (pH 7.2). To acetylate sections, tissues were exposed to 0.1 M triethanolamine, 0.25% acetic anhydride solution for 10 min. Tissues were prehybridized in hybridization buffer [50% formamide, 5× SSC, 2% blocking powder (Roche Molecular Biochemicals), 0.1% Triton X-100 (Sigma), 0.5% CHAPS (Sigma), 1 mg/ml yeast tRNA, 5 mM EDTA (pH 8.0), and 50 μg/ml heparin]. Tissues were then hybridized overnight at 55°C with the either the Prop1 or mP1 probe diluted in hybridization buffer.
The Prop1 riboprobe was generated as previously described (Cushman et al. 2001). The mP1 riboprobe was generated by subcloning the mP1 polyA region from a modified version of the pnlacF plasmid into pBluescript SK+ (Stratagene) at the BamHI and BglII sites (Peschon et al. 1987). The clone was linearized by digestion with BamHI to generate the antisense probe. The Prop1 and mP1 riboprobes were generated and labeled with digoxignenin (Roche Molecular Biochemicals) following standard procedures (Mannheim 1996).
Nontransgenic P1 pituitaries were analyzed for αGSU expression with a polyclonal rabbit anti-rat αGSU antibody (1:1800; National Institute of Diabetes and Digestive Kidney Diseases, Torrance, CA) and detected with a biotin-conjugated anti-rabbit IgG (1:400; Vector Laboratories, Burlingame, CA) using the Vectastain ABC kit (according to manufacturer’s protocol; Vector Laboratories).
PIT1 immunohistochemistry was preformed on 6-μm paraffin sections of dissected pituitary tissue as described (Charles et al. 2005).
All images were captured with a Leitz DMRB microscope (W. Nuhsbaum, Inc., McHenry, IL) and an Optronics (Goleta, CA) camera.
Results
PROP1 protein conservation
In humans, PROP1 comprises three exons that encode a 226-amino-acid protein that contains a DNA-binding homeodomain and a transactivation domain at the C-terminus. Previous studies comparing the bovine PROP1 protein sequence to that of other mammals revealed that the homeodomain is highly conserved whereas the N-terminus is not (Guy et al. 2004; Showalter et al. 2002). To obtain PROP1 protein sequences from several species for comparison, we designed primers to regions of the PROP1 gene sequence exhibiting high conservation between mouse and human. We used these primers to amplify genomic DNA from capuchin monkey, howler monkey, gorilla, and baboon. We sequenced the amplification products, aligned the genomic sequences, and compared them. The splice junctions are highly conserved, which permitted prediction of the cDNA and protein sequence (see Materials and methods).
Human PROP1 protein sequence was compared to the PROP1 protein sequence of various primate species, which are close relatives of humans, ranging from the hominoid primate clade, which shared a common ancestor with humans about 6–8 million years ago, to old-world and new-world monkeys which diverged approximately 25 and 40 million years ago, respectively (Nei et al. 2001; Nobrega and Pennacchio 2004). Human PROP1 protein sequence was also compared to the PROP1 protein sequence of more distantly related mammals. We selected the lemur (Lemur catta), which is a prosimian of intermediate evolutionary distance, having diverged from human about 60 million years ago (Boffelli et al. 2003), and more distantly related mammalian species such as artiodactyls and rodents, which are thought to have shared a common ancestor with humans over 80 million years ago (Nei et al. 2001; Nobrega and Pennacchio 2004). In addition, we compared partial protein sequences for nonmammalian vertebrates such as chicken and fish, which diverged from humans approximately 300 and 400-450 million years ago, respectively (Aparicio et al. 1995; Nei et al. 2001; Nobrega and Pennacchio 2004) (Table 1).
We compared the PROP1 protein sequence for 13 mammals via a clustalW alignment (Fig. 1). Previous PROP1 protein comparison illustrated high conservation within the homeodomain (Guy et al. 2004). In addition, two basic regions were identified within the homeodomain, B1 and B2, which are important for nuclear localization and DNA binding of PROP1 (Guy et al. 2004) (Fig. 1, boxed areas). The B1 and B2 regions are 100% identical between human, pig (Sus scrofa), cow (Bos Taurus), dog (Canis familiaris), mouse (Mus musculus), and rat (Rattus norvegicus) (Guy et al. 2004). Our analysis shows that the homeodomain is 100% conserved between humans, chimpanzees (Pan troglodytes), gorillas, and baboons (Fig. 1, shaded area and Table 1). In fact, the homeodomain region is over 93% conserved between mouse and human (Fig. 1, Table 1) and over 93% conserved between chicken (Gallus gallus) and human (Table 1). Even very distantly related vertebrate fish such as fugu (Fugu rubripes) and zebrafish (Danio rerio) show extensive conservation with human in the PROP1 homeodomain region (Table 1). Although we were surprised that the predicted fish proteins are much more divergent from the mammalian proteins than the bird (chicken), an extensive cladistic analysis of the paired domains of fish and mammalian genes support the idea that the fish sequences we present are the likely orthologs of the mammalian sequences (data not shown). In addition, all of the species that we analyzed had 100% identity in the B1 and B2 regions with the exception of the chicken, in which only partial sequence was available (data not shown), and the lemur, which had an in-frame deletion of R71 (Fig. 1, indicated by white box). This is consistent with the expectation that the homeodomain region is an important functional region of the PROP1 protein because it has evolved slowly compared to the other domains. To date all of the mutations in the human population that are known to cause MPHD as a result of PROP1 deficiency are predicted to eliminate function of the homeodomain, with the exception of the recently discovered nonsense mutation that occurs in the transactivation domain. The homeodomain mutations include various missense mutations (Fig. 1) as well as deletions, truncations, nonsense mutations, and splicing mutations (Cushman and Camper 2001; Parks et al. 1999; Reynaud et al. 2005). The homeodomain also contains the point mutation of the Ames dwarf mouse (Sornson et al. 1996) (Fig. 1). The transactivation domain is also well conserved among mammals, ranging from approximately 97%-98% identity between human and other hominoids to 76.5% identity between human and mouse (Fig. 1, Table 1). The N-terminus is the least conserved domain of PROP1, with only approximately 80% and approximately 78% identity between human and the closely related capuchin and howler primates, respectively, and less than 50% identity between human and mouse (Fig. 1).
PROP1 protein comparison reveals lack of conservation in the amino terminal domain. The PROP1 protein sequence alignment for 12 mammals shows very high conservation in the homeodomain region (shaded area) and in the transactivation domain in the carboxy terminus. The B1 and B2 regions within the homeodomain are boxed. The white box indicates the deleted amino acid within the B1 domain of the lemur. Mutations in those amino acids within the homeodomain region that are known to cause MPHD in humans (*) and mice (#) are indicated. Allelic variants in the sheep ($), cow (!), mouse (+), and human (^) are indicated. Human SNPs for PROP1 were obtained from NCBI website (http://www.ncbi.nlm.nih.gov/). Arrows indicate the exon boundaries. E1, exon 1; E2, exon 2; E3, exon 3
There are polymorphisms that occur within the PROP1 protein of humans, as well as in that of sheep, mice, and cows. (Fig. 1). Allelic variants occur in the transactivation domain in sheep (Ovis aries), T181A (Guy et al. 2004), cow, H173R (Showalter et al. 2002), and mouse, G155A, S171A, and 208/209 P insertion (Sornson et al. 1996). There is also a mouse polymorphic deletion of one CA within a CA repeat in the mouse UTR (Sornson et al. 1996). The human polymorphisms S20N, A51G, and G60E are located in the N-terminus, while A142T (Nakamura et al. 1999b), is located in the transactivation domain. The 21 additional human polymorphisms located in the noncoding sequence and silent mutations within exons 1 and 2 are listed at the SNP NCBI site (http://www.ncbi.nlm.nih.gov/). Although none of these polymorphisms have been implicated in disease, they may play a role in the genetic variation of quantitative traits within the animal kingdom.
PROP1 sequence comparison reveals three conserved noncoding elements
Previous studies using 5′ RACE located the human PROP1 transcription initiation site 309 nucleotides upstream of the translation initiation site (Duquesnoy et al. 1998). We predicted the transcription initiation site of the mouse Prop1 by comparing the mouse genomic sequence with the consensus 5′ sequence of four mouse Prop1 cDNA clones obtained from full-length cap-trapper cDNA libraries (Carninci et al. 2003). This revealed that the transcription initiation site is located 353 nucleotides upstream of the translation start site. This is 126 nucleotides upstream of the previous transcription initiation site annotated by the GenBank sequence NM008936 (http://www.ncbi.nlm.nih.gov/). Our results indicate a similar length for the human and mouse 5′ UTR, but the sequence of this region is poorly conserved (data not shown).
We compared the 5q35.3 chromosome region containing human PROP1 with the orthologous region in mouse using the UCSC genome browser (http://www.genome.ucsc.edu/), (Fig. 2A) (Kent et al. 2002). The nearest known 5′ neighboring gene to PROP1 is N4BP3, located 118 kb upstream of PROP1, and the nearest known 3′ neighboring gene, AK126616, is 34 kb downstream. In comparison, the nearest 5′ and 3′ genes to the mouse Prop1 gene are located 15 kb (Olfr1378) and 7 kb (4933414115Rik) in distance, respectively. This analysis also reveals that the mouse genomic region orthologous to the approximately 620-kb flanking region 5′ of the human PROP1 gene is inverted and separated from mouse Prop1 by a series of olfactory genes that map to human chromosomes 16 and 17 (Fig. 2A, lined box). An approximately 760-kb region located approximately 750 kb 5′ to the human PROP1 gene is present in the reverse orientation 3′ to the mouse Prop1 gene (Fig 2A, gray box). Additional comparisons of the Prop1 locus between human and chimpanzee and between rat and mouse reveal gene order conservation among primates and among rodents (data not shown). A genome VISTA comparison of the pig Prop1 BAC to the mouse genome indicated gene order conservation (Couronne et al. 2003), suggesting that the region surrounding the pig Prop1 locus is more similar to the mouse Prop1 locus than to the human (Fig. 2B). These disruptions in the gene order between the orthologous regions surrounding the human and mouse or pig PROP1 genes may obscure the identification of conserved elements located at great distances from the gene, but comparative genomics is useful for the identification of putative regulators within 26 kb that extends 5′ and 3′ from mouse Prop1 to the nearest-neighboring genes.
Mammalian genomic DNA sequence comparisons reveal the presence of several highly conserved elements. A The approximate position of BAC RP23-250122, used for transgene correction of the Prop1-deficient phenotype, is indicated at the top (double arrow), aligned with the genome sequence from the mouse. Orthologous regions of the human chromosome 5q35.3 and the mouse chromosome 11B1.3 reveal disruptions in gene order. An approximately 760-kb region that is about 750 upstream of human PROP1 is inverted and located approximately 10 kb 3′ of the mouse Prop1 (gray box, arrow). An approximately 620-kb region 5′ to the human PROP1 is inverted and located about 50 kb 5′ to the mouse Prop1 (blue box, arrow). The nearest genes 5′ and 3′ to both the human and the mouse PROP1 genes are shown. Large genes are indicated by black bars. The PROP1 gene locus is indicated by the red box and red arrow. Scale bar is marked in 175 kilobase (kb) increments. The arrow indicates the orientation of the PROP1 gene. MMu, mouse orthologs to the human 5q35.3 are indicated by a bracket and mouse chromosome number. HSa, human orthologs to the mouse 11B1.3 are indicated by a bracket and human chromosome number. B Genome VISTA plot comparing the pig Prop1 BAC to the mouse genome revealing gene order conservation. Colored peaks represent greater than 75% identity over 100 bp. The mouse Zfp354b, Prop1, and Olfr1378 genes are indicated by the arrows. C Pairwise comparisons are presented for human vs. lemur, pig, and mouse. Colored peaks represent greater than 75% identity over 100 bp. The regions of gene used for the analysis of the conserved noncoding regions are indicated by the black bars and dotted lines below the respective peaks. The PROP1 gene is indicated by the gray arrow. B, C x-axis, kilobases; y-axis, percent identity ranging from 50% to 100%. Light blue, untranslated regions (UTR); dark blue, coding regions; pink, conserved noncoding regions; bp, base pairs; CE-A, conserved noncoding element A; CE-B, conserved noncoding element B; CE-C, conserved noncoding element C (URL: http://www-gsd.lbl.gov/VISTA/)
To determine whether all of the elements necessary for appropriate regulation of Prop1 transcription are contained within a reasonably close distance to the gene, we sought to rescue the Prop1 dwarf phenotype using a mouse BAC containing Prop1. Prop1 mutants have profound, proportional dwarfism evident within the first two weeks of life, and adult mutants are approximately one third the size of their normal littermates (Buckwalter et al. 1991). Furthermore, their pituitaries have little or no GH, TSH, and PRL and exhibit anterior lobe hypoplasia and overall pituitary dysmorphology (Gage et al. 1996b; Nasonkin et al. 2004; Ward et al. 2005). The mouse BAC RP23-250I22 is about 195 kb long and contains genomic sequence extending from within the Olfr54 gene at the 5′ end of Prop1 to the Znf354c gene at the 3′ of Prop1 (Fig. 2A, double arrow). Therefore, this BAC contains additional sequence and predicted genes past the immediate-neighboring genes for Prop1. We injected this BAC into pronuclei of eggs derived from a cross of wild-type and Prop1 +/df mice. Seven transgenic lines were established, of which three were founded by Prop1 +/df mice and four by Prop1 +/+ mice. Two BAC transgenic Prop1 +/+ lines were crossed to Prop1 +/df mice to generate additional lines of BAC transgene; Prop1 +/df. These transgenic heterozygotes were crossed to Prop1 +/− mice to generate BAC transgene; Prop1 df/− mice and other assorted genotypes. Prop1 df/− mice that harbor the Prop1-containing BAC did not have a dwarf phenotype but instead were equivalent in size to Prop1 +/+and Prop1 +/− littermates (Fig. 3A, B). Of the five BAC transgenic lines tested, three produced multiple offspring (n = 2-4) in which the presence of the BAC rescued the Prop1 df/− dwarf phenotype. Among these lines there were no cases of BAC transgene; Prop1 df/− mice that were smaller than normal. One line produced multiple offspring (n = 2) in which the presence of the BAC did not rescue the Prop1 df/− dwarf phenotype, suggesting that the BAC was not intact or integrated in a region of the genome incompatible with functionally appropriate expression. One line did not produce the desired combination of the BAC transgene with Prop1 df/− in 30 pups examined, suggesting that the BAC integrated on chromosome 11.
A Prop1 BAC transgene rescues the Prop1 mutant phenotype. A Four sample mice and corresponding genotypes. Prop1 df/− mice are dwarf, but the presence of the BAC transgene rescues the dwarf phenotype. B The average weight of mice of each genotype is graphed for all progeny of a cross of BAC transgenic; Prop1 df/+ × Prop1 +/−. Error bars represent the standard deviation. For Prop1 +/+ and Prop1 +/− mice, 116 mice were weighed at weaning, while 19 and 11 were weighed for Prop1 df/− and BAC transgenic; Prop1 df/−, respectively. C Dissected pituitaries from P21 mice. The genotypes are as follows: a = Prop1 +/+, b = BAC transgenic; Prop1 df/−, c = Prop1 df/−. D-F PIT1 immunohistochemistry. The genotypes for each image are as follows: D Prop1 +/+, E BAC transgenic; Prop1 df/−, F Prop1 df /−
The BAC transgene; Prop1 df/− mice exhibiting phenotypic rescue of body size have a pituitary gland morphology indistinguishable from that of wild-type littermates, while nontransgenic Prop1 df/− mice have obviously hypoplastic anterior lobes (Fig. 3C). In addition, the BAC transgene; Prop1 df/− pituitaries contained cells that express PIT1 (Fig. 3D-F), GH, PRL, and TSH (data not shown), which are essentially absent in the Prop1 df/− dwarf pituitaries. Therefore, the Prop1-containing BAC is able to restore the pituitary cell types that are absent in the pituitaries of Prop1 df/− mice, as well as pituitary size, somatic growth, and function of pituitary target organs.
We compared a 20-kb region of the human PROP1 genomic sequence with similar portions of genomic DNA from three mammalian species: lemur, pig, and mouse. We discovered regions in the noncoding sequence of PROP1 that are highly conserved over evolutionary time (Fig. 2C). Approximately 10 kb of this sequence overlapped in all four mammals and was analyzed by the mVISTA sequence comparison program (Bray et al. 2003), which displays regions of sequence conservation between two species that are at least 100 bp long with 75% identity (Fig. 2C). The human PROP1 gene is approximately 4 kb long and is located in a region of the genome that contains relatively few genes. Exons 2 and 3, which encode the HD and TA domains (Fig. 1), are highly conserved among all four species (Fig. 2C, blue peaks). Exon 1, however, is relatively divergent among these species (Fig. 2C). The mVISTA plot also reveals three conserved noncoding sequences among human, lemur, pig, and mouse (Fig. 2C, pink peaks). The first conserved element A (CE-A) is located in the 5′ flanking sequence 9 bp upstream of the mouse transcription initiation site and is 80.5% identical between human and mouse over 312 bp (Fig. 2C). CE-B is located within intron 1 of Prop1 and is 81.8% identical between human and mouse over 176 bp (Fig. 2C). The third conserved region, CE-C, is the smallest conserved noncoding element and is only 75.7% identical between human and mouse with a length of approximately 100 bp (Fig. 2C).
The Prop1 CE-A and CE-B regions show enhancer activity in tissue culture
Various in vitro and in vivo methods have been used to test the biological function of conserved elements discovered by comparative genomics (Boffelli et al. 2003; Lettice et al. 2003; Nobrega et al. 2003; Zerucha et al. 2000). CE-A has no promoter activity in a heterologous CV1 monkey kidney or the αT3 mouse gonadotrope-like cell lines (data not shown). Therefore, CE-A, CE-B, and CE-C were analyzed for enhancer activity in tissue culture by inserting each of them in both the forward and the reverse orientation upstream of the Drosophila alcohol dehydrogenase (ADH) minimal promoter driving a luciferase (Luc) reporter gene (Fig. 4A, construct #1). The cell lines used for the transfection assays included the CV-1, αT3, GH3 rat somatotrope-like, and AtT-20 mouse corticotrope-like cell lines. The enhancer activity of these elements is reported as the fold increase in luciferase activity over the basal level of the ADH-Luc vector alone.
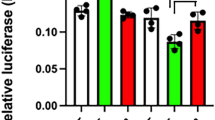
Enhancer study of Prop1 conserved noncoding elements from the mouse gene in monkey kidney CV-1, GH3 (rat somato-lactotropes), αT3-1 (mouse gonadotrope-like), and AtT-20 (mouse corticotrope-like) cells. A Diagram of the conserved elements upstream of the ADH-Luc reporter construct. B Luciferase activity as fold activation over basal shown for the constructs depicted in (A) for CV-1, GH3, αT3-1, and AtT-20 cells reveal orientation-dependent enhancer activity for CE-A in all lines and for CE-B in CV-1 and GH3 lines. ADH, alcohol dehydrogenase minimal promoter; LUC, luciferase reporter gene; F, forward orientation; R, reverse orientation. The asterisk designates constructs with activity significantly above baseline
A 584-bp Prop1 5′ flanking segment containing the CE-A element from the mouse Prop1 gene was cloned into the ADH-Luc vector in both forward and reverse orientations (Figs. 2C and 4A, constructs #2 and #3, respectively). On average, the CE-A (F)/ADH construct displayed about a 13.5-fold increase in activity in the CV-1 cells and approximately 4-5-fold increase in activity in the GH3, αT3-1, and AtT-20 cell lines (Fig. 4B). When the CE-A was tested in the reverse orientation (Fig. 4A, construct #3), the CE-A (R)/ADH showed no activity in any of the cell lines tested, indicating that the effect is orientation-dependent (Fig. 4B).
A 508-bp segment from intron 1 of the mouse Prop1 gene containing the CE-B element was also tested for enhancer activity in the ADH-Luc reporter construct (Figs. 2C and 4A, constructs #4 and #5, respectively). The CE-B (F)/ADH construct elicited, on average, a fourfold increase in luciferase activity compared to the ADH-Luc alone in the CV-1 cells (Fig. 4B). However, the CE-B (F)/ADH had very low activity in the GH3 cells (less than a twofold average increase over basal with a range of 1.1–2.6-fold) and no activity in either the αT3-1 or the AtT-20 lines (Fig. 4B). The CE-B (R)/ADH construct showed no enhancer activity in any of the cell lines tested (Fig. 4B).
A 1196-bp segment from the 3′ flanking sequence of the mouse Prop1 gene containing CE-C was inserted into the ADH-Luc reporter construct (Figs. 2C and 4A, constructs #6 and #7, respectively). A larger section was used in the analysis of the CE-C element because there were several smaller peaks on the mVISTA plot between human and mouse that showed significant conservation between human and pig and between human and lemur that flanked the CE-C segment (Fig. 2C). The CE-C in either orientation appeared to have no enhancer activity in the αT3-1 or the GH3 cell lines but did have a low level of activity in the forward orientation when tested in the CV-1 and AtT-20 cell lines (Fig. 4B).
In summary, the 5′ element CE-A had the highest activity with the ADH-Luc reporter. The CE-B intronic element and the CE-C 3′ element had low orientation-dependent enhancer activity in the CV-1 cells and very low, if any, activity in the pituitary-derived cell lines. This was not surprising because these pituitary cell types are derived from differentiated pituitary cells and do not express endogenous Prop1. These pituitary-like cells may not have all the components necessary to allow for the proper function of all of the Prop1 enhancer elements or they may contain factors that repress the function of these elements.
The Prop1 CE-A element is not sufficient for LacZ expression in transgenic mice
The CE-A and CE-B regions were also analyzed in transgenic mice to determine whether they function in vivo. CE-A/LacZ (Table 2), which consists of a 3-kb region immediately 5′ to the Prop1 ATG containing the 5′ UTR, the Prop1 transcription initiation site, and the CE-A region, was analyzed for the ability to drive the expression of a nuclear localized E. coli β-galactosidase-mouse protamine 1 reporter construct (LacZ-pA) in transgenic mice (Fig. 2C). Transgenic founders were bred to produce lines for embryo analysis, or transient transgenic embryos were harvested and analyzed for the transgenic transcript via in situ hybridization (ISH) at e12.5 because this is the time of peak Prop1 expression (Sornson et al. 1996). We used the well-characterized αGsu-LacZ and αGsu-Prop1 transgenes as positive controls for detecting β-galactosidase activity in the pituitary gland by X-gal staining and for mouse protamine ISH, respectively (Cushman et al. 2001; Kendall et al. 1994). The promoter and enhancer sequences of the pituitary glycoprotein hormone α-subunit gene (Cga for chorionic subunit α or αGsu) are sufficient to drive expression of LacZ and other reporters in a manner that is developmentally, hormonally, and tissue-specifically correct (Kendall et al. 1994). Both the αGsu-LacZ transgenic mice and the αGsu-Prop1 transgenic mice contain the mouse protamine splice sites and polyadenylylation sequences that can be used as a transgene-specific tag in any organ except the testis, which expresses the endogenous protamine gene. None of the five transgenic embryos exhibited detectable transcription of the CE-A/LacZ transgene compared with positive controls. Thus, the 3-kb 5′ flanking region of Prop1 is not sufficient for transgene expression. The intronic CE-B element was tested in conjunction with CE-A for in vivo Prop1 expression. Transgenic mice were made with the construct CE-B+CE-A/LacZ, which contains CE-B upstream of the Prop1 3-kb 5′ flanking region (Fig. 2C, Table 2). Fifteen transient transgenic embryos were harvested at e12.5 and assayed for expression of the transgene by either X-gal staining or ISH for mP1. None of the 15 embryos analyzed had detectable expression of the transgene, although the positive controls did exhibit expression. Thus, the combination of CE-B and CE-A is also not sufficient for expression in vivo.
The Prop1 CE-B putative enhancer is important for spatial expression in transgenic mice
The CE-B region was tested for enhancer function in transgenic mice in the context of the αGsu promoter. The first clues to a possible role for CE-B in the expression of Prop1 came from the analysis of the αGsu-Prop1 transgenic mice (Table 2). Endogenous Prop1 is normally expressed from e9 until e15.5 in the mouse in a dorsal (highest) to ventral (lowest) expression gradient (Fig. 5A) (Sornson et al. 1996). In contrast, αGsu is expressed during pituitary development in the caudomedial and ventral portions of the pituitary where the thyrotropes and gonadotropes arise, and its expression continues in these two cell types throughout adulthood (Fig. 5B) (Japon et al. 1994; Kendall et al., 1994; Raetzman et al. 2002). Ten stable lines were created with the αGsu-Prop1 transgene and six expressed the transgene in the adult pituitary. Two lines were maintained and analyzed (Cushman et al. 2001). These studies revealed that the αGsu-Prop1 construct yields efficient overexpression of Prop1 beyond e15.5 (Cushman et al. 2001). Expression of the αGsu-Prop1 transgene is expanded dorsally beyond the normal expression domain of endogenous αGsu and beyond that observed for other αGsu driving expression of LacZ or other reporters. This dorsal expression is observed in pituitaries at postnatal day 1 (P1, day of birth) in 2/2 stable transgenic lines and at e18.5 in 3/3 transient transgenics that were created with the same αGsu-Prop1 construct. This expression pattern appears to be a combination of both endogenous αGsu and Prop1 (compare Fig. 5C with B and A). This result suggests that the CE-B region is important for the dorsal expansion of the αGsu-Prop1 transgene and thus important for the spatial expression of endogenous Prop1.
Transgenic mice reveal functional properties of CE-B. A Endogenous Prop1 expression illustrated diagrammatically and with Prop1 in situ hybridization in sagittal e12.5 and e14.5 mouse and coronal e14.5 pituitaries. A dorsal-to-ventral expression gradient is apparent. The anterior lobe is marked by a black line. B Immunohistochemical staining with αGSU antibody (brown DAB chromagen) reveals normal αGsu expression in the caudomedial and ventral regions of a P1 coronal pituitary expression via IHC. C In situ hybridization for Prop1 transcripts [purple AP chromagen] reveals that the αGsu-Prop1 transgenic construct expression is expanded dorsally in P1 coronal pituitary sections relative to endogenous αGsu expression. D Prop1 in situ hybridization shows that the αGsu-Prop1ΔCE-B transgenic construct expression (purple AP chromagen) was restored to the caudomedial and ventral regions in P1 coronal pituitaries. E Prop1 in situ hybridization shows that the CE-B + αGsu Prop1ΔCE-B transgenic expression (purple AP chromagen) was expanded to the dorsal aspects of the pituitary at P1. C–E Representations of the transgenic constructs are shown beneath the appropriate studies. Brackets mark the region for each pituitary in which the transgene expression was detected. F In situ hybridization for Prop1 in a nontransgenic pituitary gives no signal. DAB, 3,3′ diaminobenzidine; AP, alkaline phosphatase; A, anterior lobe; P, posterior lobe; I, intermediate lobe; RP, Rathke’s pouch; INF, infindibulum; v, ventral expression pattern; d + v, dorsal and ventral expression pattern
We tested the significance of CE-B for dorsalized transgene expression by deleting CE-B from intron 1 to produce the construct αGsu-Prop1ΔCE-B (Table 2). Transient transgenics bearing this construct were harvested at P1 and assayed for the expression of the transgene using ISH for Prop1. Six of the eight transgenic mice analyzed exhibited transgene expression. One had very weak expression and was eliminated from further consideration. The remaining five had strong expression. In these five transgenic mice, the expression pattern of the αGsu-Prop1ΔCE-B transgene is restricted to the ventral aspects of the pituitary, typical for the αGsu promoter and enhancer (compare Fig. 5D with C). This result indicates that CE-B was necessary for the dorsal expression of the αGsu-Prop1 construct.
To determine whether the position of the CE-B relative to the αGsu promoter is critical, the CE-B region was replaced in the αGsu-Prop1ΔCE-B construct upstream of the αGsu promoter to create a new construct, CE-B + αGsu-Prop1ΔCE-B (Table 2). The CE-B + αGsu-Prop1ΔCE-B transgenic mice were harvested at P1 and assayed for expression of the transgene. The CE-B + αGsu-Prop1ΔCE-B transgene expression was expanded to the dorsal aspect of the pituitary in six of seven transient lines (Fig. 5E), with the remaining line having only weak expression of the transgene. Finally, an ISH for Prop1 on nontransgenic pituitaries gave no signal (Fig. 5F), thus verifying that the patterns of expression seen in Figs. 5C, D, F are due to the specific transgene. The transgenic analysis provided in vivo evidence that the CE-B contained within the intron 1 of Prop1 is sufficient to confer spatial expression information in a position-independent manner in the context of the transgene.
Discussion
We sought to identify cis-acting DNA sequences important for mouse Prop1 expression because regulation of Prop1 is important for normal pituitary development and function. To accomplish this, we obtained genomic sequence from lemur and pig (Sus scrofa) PROP1 BACs and compared these to PROP1 sequences available online (Ahituv et al. 2004; Aparicio et al. 1995; Boffelli et al. 2003, 2004; Nobrega and Pennacchio 2004; Williams et al. 2003). We also generated PROP1 genomic sequence for the first time for five different primate species to include in the comparison. We identified three conserved noncoding elements (CE) that are larger than 100 bp with greater than 75% identity between human and mouse and tested them for function in cell culture and transgenic mice. The three regions that fit these criteria are CE-A, a 300-bp region in the Prop1 promoter proximal region; CE-B, a 200-bp region within the first intron of Prop1; and CE-C, a 103-bp region within the 3′ flanking sequence.
Transfection of cultured cells has been successful for demonstrating the function of some elements (Nishimura et al. 2000; Surinya et al. 1998; Swamynathan and Piatigorsky 2002), but there are examples of important regulatory sequences that are not identified with this approach (Lang et al. 2003; Lettice et al. 2003; Nobrega et al. 2003; Zerucha et al. 2000). Prop1 is expressed in a distinct spatial-, temporal-, and tissue-specific fashion during development, and the endogenous gene is not expressed in any of the available pituitary cell lines. Nevertheless, CE-A exhibited orientation-dependent activity in all cell lines. CE-B, located within intron 1 of Prop1, also appeared to have orientation-dependent enhancer activity in CV-1 cells and GH3 cells, although at a much lower level. There was no enhancer activity in either the AtT-20 line or the αT3-1 line. The CE-C putative regulatory element had low enhancer activity in only the CV-1 and AtT-20 cell lines. There are many possible explanations for the weak activity of CE-B and CE-C in cell culture, but the cell culture assays did detect enhancer function. The pituitary-derived cell lines were developed in the different hormone-producing cell lineages, e.g., the αT3-1 cells are gonadotrope-like, whereas GH3 cells are somatotrope-like. Therefore, the differential enhancer activities of the different constructs in these cell lines may be examples of context-specific activity. In addition, the cell lines may be more representative of differentiated cell types and, because Prop1 is expressed significantly only during early pituitary development in the rodent (Sornson et al., 1996), these cells may not contain the transcription factors and cofactors necessary for the full activity of the putative enhancers. These enhancers also are orientation-specific. However, other examples of orientation-dependent enhancers have been reported (Cheng et al. 2004; Falvo et al. 2000; Nishimura et al. 2000; Surinya et al. 1998; Swamynathan and Piatigorsky 2002; Wei and Brennan 2000).
Our transgenic experiments shed some light on the function of the CE-B region. In the context of the αGsu promoter, the CE-B element results in dorsal expansion of transgene expression. Although this construct used a heterologous promoter, which allows for the expression of the transgene after the endogenous Prop1 expression is extinguished, these results provide evidence that the CE-B region in intron 1 of Prop1 is important for spatial expression. The CE-B region in conjunction with the αGsu promoter will be useful for driving the expression of transgenes in the more dorsal aspects of the developing pituitary. Other studies have shown that the Rbp-Jκ DNA binding protein, which is the primary mediator of Notch signaling, can directly bind to intron 1 of Prop1 and is important for the maintenance of Prop1 expression (Zhu et al. 2006). Taken together, these data suggest an in vivo role for the CE-B in the regulation of Prop1 expression.
Two kilobase pairs of the Prop1 promoter proximal region (CE-A) is inadequate for reporter gene expression in transgenic mice, even in the context of CE-B. This indicates that additional sequences are necessary for Prop1 expression in mice. The BAC rescue of Prop1 −/df mice demonstrates that all of the elements necessary for transcriptional regulation of Prop1 are contained within the BAC. We predict that the remaining critical sequences for Prop1 expression are within the region immediately surrounding Prop1, within 15 kb upstream and approximately 26 kb downstream, because there is a disruption in gene order between human and mouse or pig. These critical control sequences are not readily identifiable by genomic sequence comparisons.
In summary, we identified a region, contained within intron 1 of Prop1, which is necessary and sufficient for the spatial expression of Prop1 in the context of a heterologous pituitary specific promoter. While additional regulatory elements remain to be identified by other approaches, the intronic element is worth screening for mutations in unexplained cases of MPHD patients, especially those that appear heterozygous for mutations in PROP1.
References
Agarwal G, Bhatia V, Cook S, Thomas PQ (2000) Adrenocorticotropin deficiency in combined pituitary hormone deficiency patients homozygous for a novel PROP1 deletion. J Clin Endocrinol Metab 85:4556–4561
Ahituv N, Rubin EM, Nobrega MA (2004) Exploiting human–fish genome comparisons for deciphering gene regulation. Hum Mol Genet 13 Spec No 2:R261–R266
Aparicio S, Morrison A, Gould A, Gilthorpe J, Chaudhuri C, et al. (1995) Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc Natl Acad Sci U S A 92:1684–1688
Bagheri-Fam S, Ferraz C, Demaille J, Scherer G, Pfeifer D (2001) Comparative genomics of the SOX9 region in human and Fugu rubripes: conservation of short regulatory sequence elements within large intergenic regions. Genomics 78:73–82
Bhangoo AP, Hunter CS, Savage JJ, Anhalt H, Pavlakis S, et al. (2006) Clinical case seminar: a novel LHX3 mutation presenting as combined pituitary hormonal deficiency. J Clin Endocrinol Metab 91:747–753
Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovcharenko I, et al. (2003) Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299:1391–1394
Boffelli D, Nobrega MA, Rubin EM (2004) Comparative genomics at the vertebrate extremes. Nat Rev Genet 5:456–465
Bottner A, Keller E, Kratzsch J, Stobbe H, Weigel JF, et al. (2004) PROP1 mutations cause progressive deterioration of anterior pituitary function including adrenal insufficiency: a longitudinal analysis. J Clin Endocrinol Metab 89:5256–5265
Braga S, Phillips JA 3rd, Joss E, Schwarz H, Zuppinger K (1986) Familial growth hormone deficiency resulting from a 7.6 kb deletion within the growth hormone gene cluster. Am J Med Genet 25:443–452
Bray N, Dubchak I, Pachter L (2003) AVID: A global alignment program. Genome Res 13:97–102
Brinkmeier ML, Potok MA, Cha KB, Gridley T, Stifani S, et al. (2003) TCF and Groucho-related genes influence pituitary growth and development. Mol Endocrinol 17:2152–2161
Buckwalter MS, Katz RW, Camper SA (1991) Localization of the panhypopituitary dwarf mutation (df) on mouse chromosome 11 in an intersubspecific backcross. Genomics 10:515–526
Carninci P, Waki K, Shiraki T, Konno H, Shibata K, et al. (2003) Targeting a complex transcriptome: the construction of the mouse full-length cDNA encyclopedia. Genome Res 13:1273–1289
Charles MA, Suh H, Hjalt TA, Drouin J, Camper SA, et al. (2005) PITX genes are required for cell survival and Lhx3 activation. Mol Endocrinol 19:1893–1903
Cheng HC, Wang CK, Upholt WB (2004) Transcriptional regulation of Msx2 in the AERs of developing limbs is dependent on multiple closely spaced regulatory elements. Dev Biol 270:513–524
Cogan J, Wu W, Phillips JI, Arnhold I, Agapito A, et al. (1998) The PROP1 2-base pair deletion is a common cause of combined pituitary hormone deficiency. J Clin Endocrinol Metab 83:3346–3349
Cohen LE, Wondisford FE, Radovick S (1996) Role of Pit-1 in the gene expression of growth hormone, prolactin, and thyrotropin. Endocrinol Metab Clin North Am 25:523–540
Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, et al. (2003) Strategies and tools for whole-genome alignments. Genome Res 13:7
Cushman LJ, Camper SA (2001) Molecular basis of pituitary dysfunction in mouse and human. Mamm Genome 12:485–494
Cushman LJ, Watkins-Chow DE, Brinkmeier ML, Raetzman LT, Radak AL, et al. (2001) Persistent Prop1 expression delays gonadotrope differentiation and enhances pituitary tumor susceptibility. Hum Mol Genet 10:1141–1153
Dasen JS, Barbera JP, Herman TS, Connell SO, Olson L, et al. (2001) Temporal regulation of a paired-like homeodomain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis. Genes Dev 15:3193–3207
Deladoey J, Fluck C, Buyukgebiz A, Kuhlmann BV, Eble A, et al. (1999) “Hot spot” in the PROP1 gene responsible for combined pituitary hormone deficiency. J Clin Endocrinol Metab 84:1645–1650
Duquesnoy P, Roy A, Dastot F, Ghali I, Teinturier C, et al. (1998) Human Prop-1: cloning, mapping, genomic structure. Mutations in familial combined pituitary hormone deficiency. FEBS Lett 437:216–220
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Falvo JV, Parekh BS, Lin CH, Fraenkel E, Maniatis T (2000) Assembly of a functional beta interferon enhanceosome is dependent on ATF-2-c-jun heterodimer orientation. Mol Cell Biol 20:4814–4825
Fluck C, Deladoey J, Rutishauser K, Eble A, Marti U, et al. (1998) Phenotypic variability in familial combined pituitary hormone deficiency caused by a PROP1 gene mutation resulting in the substitution of Arg→Cys at codon 120 (R120C). J Clin Endocrinol Metab 83:3727–3734
Gage PJ, Brinkmeier ML, Scarlett LM, Knapp LT, Camper SA, et al. (1996a) The Ames dwarf gene, df, is required early in pituitary ontogeny for the extinction of Rpx transcription and initiation of lineage specific cell proliferation. Mol Endocrinol 10:1570–1581
Gage PJ, Roller ML, Saunders TL, Scarlett LM, Camper SA (1996b) Anterior pituitary cells defective in the cell-autonomous factor, df, undergo cell lineage specification but not expansion. Development 122:151–160
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Guy JC, Hunter CS, Showalter AD, Smith TP, Charoonpatrapong K, et al. (2004) Conserved amino acid sequences confer nuclear localization upon the Prophet of Pit-1 pituitary transcription factor protein. Gene 336:263–273
Iniguez-Lluhi JA, Lou DY, Yamamoto KR (1997) Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J Biol Chem 272:4149–4156
Japon MA, Rubinstein M, Low MJ (1994) In situ hybridization analysis of anterior pituitary hormone gene expression during fetal mouse development. J Histochem Cytochem 42:1117–1125
Kendall SK, Gordon DF, Birkmeier TS, Petrey D, Sarapura VD, et al. (1994) Enhancer-mediated high level expression of mouse pituitary glycoprotein hormone alpha-subunit transgene in thyrotropes, gonadotropes, and developing pituitary gland. Mol Endocrinol 8:1420–1433
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656–664
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, et al. (2002) The human genome browser at UCSC. Genome Res 12:996–1006
Kleinjan DA, van Heyningen V (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76:8–32
Komminoth P (1996) Detection of mRNA in tissue sections using DIG-labeled RNA and oligonucleotide probes. In: Nonradioactive In Situ hybridization application manual, 2nd edn. Mannheim, Boehringer Mannheim, Germany, pp 126–135
Lang D, Brown CB, Milewski R, Jiang YQ, Lu MM, et al. (2003) Distinct enhancers regulate neural expression of Pax7. Genomics 82:553–560
Laumonnier F, Ronce N, Hamel BC, Thomas P, Lespinasse J, et al. (2002) Transcription factor SOX3 is involved in X-linked mental retardation with growth hormone deficiency. Am J Hum Genet 71:1450–1455
Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, et al. (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12:1725–1735
Machinis K, Pantel J, Netchine I, Leger J, Camand OJ, et al. (2001) Syndromic short stature in patients with a germline mutation in the LIM homeobox LHX4. Am J Hum Genet 69:961–968
Matsumoto A, Ishii S (eds) (1987) Atlas of Endocrine Organs (Tokyo: Springer-Verlag)
Mendonca B, Osorio M, Latronico A, Estefan V, Lo L, et al. (1999). Longitudinal hormonal and pituitary imaging changes in two females with combined pituitary hormone deficiency due to deletion of A301,G302 in the PROP1 gene. J Clin Endocrinol Metab 84:942–945
Mullis P, Patel M, Brickell PM, Brook CG (1990) Isolated growth hormone deficiency: analysis of the growth hormone (GH)-releasing hormone gene and the GH gene cluster. J Clin Endocrinol Metab 70:187–191
Nakamura S, Ohtsuru A, Takamura N, Kitange G, Tokunaga Y, et al. (1999a) Prop-1 gene expression in human pituitary tumors. J Clin Endocrinol Metab 84:2581–2584
Nakamura Y, Usui T, Mizuta H, Murabe H, Muro S, et al. (1999b) Characterization of Prophet of Pit-1 gene expression in normal pituitary and pituitary adenomas in humans. J Clin Endocrinol Metab 84:1414–1419
Nasonkin IO, Ward RD, Raetzman LT, Seasholtz AF, Saunders TL, et al. (2004) Pituitary hypoplasia and respiratory distress syndrome in Prop1 knockout mice. Hum Mol Genet 13:2727–2735
Nei M, Xu P, Glazko G (2001) Estimation of divergence times from multiprotein sequences for a few mammalian species and several distantly related organisms. Proc Natl Acad Sci U S A 98:2497–2502
Netchine I, Sobrier ML, Krude H, Schnabel D, Maghnie M, et al. (2000) Mutations in LHX3 result in a new syndrome revealed by combined pituitary hormone deficiency. Nat Genet 25:182–186
Nishimura S, Takahashi S, Kuroha T, Suwabe N, Nagasawa T, et al. (2000) A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Mol Cell Biol 20:713–723
Nobrega MA, Pennacchio LA (2004) Comparative genomic analysis as a tool for biological discovery. J Physiol 554:31–39
Nobrega MA, Ovcharenko I, Afzal V, Rubin EM (2003) Scanning human gene deserts for long-range enhancers. Science 302:413
Parks JS, Brown MR, Hurley DL, Phelps CJ, Wajnrajch MP (1999) Heritable disorders of pituitary development. J Clin Endocrinol Metab 84:4362–4370
Pennacchio LA, Rubin EM (2001) Genomic strategies to identify mammalian regulatory sequences. Nat Rev Genet 2:100–109
Peschon JJ, Behringer RR, Brinster RL, Palmiter RD (1987) Spermatid-specific expression of protamine 1 in transgenic mice. Proc Natl Acad Sci U S A 84:5316–5319
Pfäffle RW, DiMattia GE, Parks JS, Brown MR, Wit JM, et al. (1992) Mutation of the POU-specific domain of Pit-1 and hypopituitarism without pituitary hypoplasia. Science 257:1118–1121
Plessy C, Dickmeis T, Chalmel F, Strahle U (2005) Enhancer sequence conservation between vertebrates is favoured in developmental regulator genes. Trends Genet 21:207–210
Procter AM, Phillips JA 3rd, Cooper DN (1998) The molecular genetics of growth hormone deficiency. Hum Genet 103:255–272
Radovick S, Nations M, Du Y, Berg LA, Weintraub BD, et al. (1992) A mutation in the POU-Homeodomain of Pit-1 responsible for combined pituitary hormone deficiency. Science 257:1115–1117
Raetzman LT, Ward R, Camper SA (2002) Lhx4 and Prop1 are required for cell survival and expansion of the pituitary primordia. Development 129:4229–4239
Raetzman LT, Ross SA, Cook S, Dunwoodie SL, Camper SA, et al. (2004) Developmental regulation of Notch signaling genes in the embryonic pituitary: Prop1 deficiency affects Notch2 expression. Dev Biol 265:329–340
Reynaud R, Barlier A, Vallette-Kasic S, Saveanu A, Guillet MP, et al. (2005) An uncommon phenotype with familial central hypogonadism caused by a novel PROP1 gene mutant truncated in the transactivation domain. J Clin Endocrinol Metab 90:4880–4887
Showalter AD, Smith TP, Bennett GL, Sloop KW, Whitsett JA, et al. (2002) Differential conservation of transcriptional domains of mammalian Prophet of Pit-1 proteins revealed by structural studies of the bovine gene and comparative functional analysis of the protein. Gene 291:211–221
Skelly RH, Korbonits M, Grossman A, Besser GM, Monson JP, et al. (2000) Expression of the pituitary transcription factor Ptx-1, but not that of the trans-activating factor prop-1, is reduced in human corticotroph adenomas and is associated with decreased alpha-subunit secretion. J Clin Endocrinol Metab 85:2537–2542
Sloop KW, McCutchan Schiller A, Smith TP, Blanton JR Jr, Rohrer GA, et al. (2000) Biochemical and genetic characterization of the porcine Prophet of Pit-1 pituitary transcription factor. Mol Cell Endocrinol 168:77–87
Sornson MW, Wu W, Dasen JS, Flynn SE, Norman DJ, et al. (1996) Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature 384:327–333
Suh H, Gage PJ, Drouin J, Camper SA (2002) Pitx2 is required at multiple stages of pituitary organogenesis: pituitary primordium formation and cell specification. Development 129:329–337
Surinya KH, Cox TC, May BK (1998) Identification and characterization of a conserved erythroid-specific enhancer located in intron 8 of the human 5-aminolevulinate synthase 2 gene. J Biol Chem 273:16798–16809
Swamynathan SK, Piatigorsky J (2002) Orientation-dependent influence of an intergenic enhancer on the promoter activity of the divergently transcribed mouse Shsp/alpha B-crystallin and Mkbp/HspB2 genes. J Biol Chem 277:49700–49706
Szeto DP, Rodriguez-Estaban C, Ryan AK, O’Connell SM, Liu F, et al. (1999) Role of Bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev 13:484–494
Tajima T, Hattorri T, Nakajima T, Okuhara K, Sato K, et al. (2003) Sporadic heterozygous frameshift mutation of HESX1 causing pituitary and optic nerve hypoplasia and combined pituitary hormone deficiency in a Japanese patient. J Clin Endocrinol Metab 88:45–50
Tatsumi K, Miyai K, Notomi T, Kaibe K, Amino N, et al. (1992) Cretinism with combined hormone deficiency caused by a mutation in the PIT1 gene. Nat Genet 1:56–58
Usui T, Nakamura Y, Mizuta H, Murabe H, Muro S, et al. (2000) Functional significance of prop-1 gene expression in pituitary adenomas. Endocr J 47 Suppl:S85–S89
Vesper AH, Raetzman LT, Camper SA (2006) Role of prophet of Pit1 (PROP1) in gonadotrope differentiation and puberty. Endocrinology 147:1654–1663
Vimpani GV, Vimpani AF, Lidgard GP, Cameron EHD, Farquhar JW (1977) Prevalence of severe growth hormone deficiency. Br Med J 2:427–430
Ward RD, Raetzman LT, Suh H, Stone BM, Nasonkin IO, et al. (2005) Role of PROP1 in pituitary gland growth. Mol Endocrinol 19:698–710
Wei W, Brennan MD (2000) Polarity of transcriptional enhancement revealed by an insulator element. Proc Natl Acad Sci U S A 97:14518–14523
Williams SH, Mouchel N, Harris A (2003) A comparative genomic analysis of the cow, pig, and human CFTR genes identifies potential intronic regulatory elements. Genomics 81:628–639
Wu W, Cogan JD, Pfaffle RW, Dasen JS, Frisch H, et al. (1998) Mutations in PROP1 cause familial combined pituitary hormone deficiency. Nat Genet 18:147–149
Zerucha T, Stuhmer T, Hatch G, Park BK, Long Q, et al. (2000) A highly conserved enhancer in the Dlx5/Dlx6 intergenic region is the site of cross-regulatory interactions between Dlx genes in the embryonic forebrain. J Neurosci 20:709–721
Zhu X, Zhang J, Tollkuhn J, Ohsawa R, Bresnick EH, et al. (2006) Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis. Genes Dev 20:2739–2753
Acknowledgments
This work was funded by the NIH (T32 GM07863 and T32 GM07315 to RDW, NRSA F32 DK60306 to LTR, R37HD30428 and R01HD34283 to SAC), the Howard Hughes Medical Institute, and the University of Michigan Bioinformatics Program. The authors thank the University of Michigan Transgenic Animal Model Core and their supporting entities: NIH grants (CA46592, AR20557, DK07367), the University of Michigan Center for Organogenesis, the Michigan Economic Development Corporation, and the Michigan Technology Tri-Corridor (Michigan Animal Models Consortium grant 085P1000815). They thank Igor Nasonkin, Frank Probst, and Hoonkyo Suh for their help in performing the initial ClustalW alignments and consensus binding site screenings; Deborah Gumucio and Morris Goodman for kindly providing the primate genomic DNAs; Pam Mellon for providing the αT3-1 cell lines; Jorge Iniquez for the generous gift of the pDODLO 02 ADH luciferase reporter construct; and Audrey Seasholtz and Sam Holmstrom for their helpful advice.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Ward, R.D., Davis, S.W., Cho, M. et al. Comparative genomics reveals functional transcriptional control sequences in the Prop1 gene. Mamm Genome 18, 521–537 (2007). https://doi.org/10.1007/s00335-007-9008-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-007-9008-6