Abstract
The oculocerebrorenal syndrome of Lowe (OCRL; MIM #309000) is an X-linked human disorder characterized by congenital cataracts, mental retardation, and renal proximal tubular dysfunction caused by loss-of-function mutations in the OCRL gene that encodes Ocrl, a type II phosphatidylinositol bisphosphate (PtdIns4,5P2) 5-phosphatase. In contrast, mice with complete loss-of-function of the highly homologous ortholog Ocrl have no detectable renal, ophthalmological, or central nervous system abnormalities. We inferred that the disparate phenotype between Ocrl-deficient humans and mice was likely due to differences in how the two species compensate for loss of the Ocrl enzyme. We therefore turned our attention to Inpp5b, another type II PtdIns4,5P2 5-phosphatase encoded by Inpp5b in mice and INPP5B in humans, as potential compensating genes in the two species, because Inpp5b/INPP5B are the most highly conserved paralogs to Ocrl/OCRL in the respective genomes of both species and Inpp5b demonstrates functional overlap with Ocrl in mice in vivo. We used in silico sequence analysis, reverse-transcription PCR, quantitative PCR, and transient transfection assays of promoter function to define splice-site usage and the function of an internal promoter in mouse Inpp5b versus human INPP5B. We found mouse Inpp5b and human INPP5B differ in their transcription, splicing, and primary amino acid sequence. These observations form the foundation for analyzing the functional basis for the difference in how Inpp5b and INPP5B compensate for loss of Ocrl function and, by providing insight into the cellular roles of Ocrl and Inpp5b, aid in the development of a model system in which to study Lowe syndrome.
Similar content being viewed by others
Introduction
The oculocerebrorenal syndrome of Lowe (OCRL; MIM #309000) is an X-linked human disorder characterized by congenital cataracts, mental retardation, and renal proximal tubular dysfunction (Bockenhauer et al. 2008; Charnas et al. 1991; Kenworthy and Charnas 1995; Kenworthy et al. 1993; Suchy and Nussbaum 2009). OCRL is caused by loss-of-function mutations in the OCRL gene (Attree et al. 1992; Leahey et al. 1993; Lin et al. 1998; Monnier et al. 2000), which encodes Ocrl, a type II phosphatidylinositol bisphosphate (PtdIns4,5P2) 5-phosphatase (Suchy et al. 1995; Zhang et al. 1995). A previous attempt to create a mouse model for OCRL failed when mice with a complete loss-of-function mutation in Ocrl had no discernible renal, ophthalmological, or central nervous system abnormalities (Janne et al. 1998). The reference protein sequences for the Ocrl enzyme from human (NP_000267) and mouse (NP_796189) are 91% identical and 95% conserved, and the gene in both species is highly expressed in tissues relevant to OCRL, such as the brain and kidney (Janne et al. 1998) (GeneCards: www.genecards.org; SOURCE_Gene_Report: http://genome-www5.Stanford.edu). We inferred that the difference in phenotype between Ocrl-deficient humans and mice is likely not the result of a divergence in the function of the OCRL/Ocrl orthologs themselves but rather is due to differences in how the two species compensate for loss of the enzyme. Inpp5b, another type II PtdIns4,5P2 5-phosphatase (encoded by INPP5B in humans and Inpp5b in mice), is the most highly conserved paralog to Ocrl in the genomes of both species and has functional overlap with Ocrl in mice in vivo (Bernard and Nussbaum 2010; Janne et al. 1998). If the divergent phenotype in Ocrl-deficient mice and humans could be ascribed to a difference in how well Inpp5b and INPP5B compensate for loss of Ocrl function, then there should be differences in the expression and/or primary structure of the two orthologs in mice and humans. In this article we show such differences do exist. We describe a distinctive pattern of splicing of one exon (exon 7) and measure a quantitative difference between the two species in the activity of an internal promoter near exon 7.
Materials and methods
Northern blot analysis
Northern blot analysis was performed using Clontech commercial mouse blots, hybridized per the manufacturer’s instructions (Clontech Laboratories, Inc., Mountain View, CA).
RT-PCR of mouse RNA
For mouse RNA, brain and kidneys were dissected from mice and stored at 4°C overnight in RNAlater (Ambion, Austin, TX). RNA was isolated from tissues using Trizol (Invitrogen, Carlsbad, CA). Human RNA used was human brain total RNA (Ambion AM7962) and human kidney total RNA (Ambion AM7976). RNA was converted to cDNA using a First Strand cDNA Synthesis kit with random primers (GE Healthcare 27-9261-01) according to the manufacturer’s instructions. Once cDNA was made, 1 μl of this reaction mixture was used in a standard PCR reaction using forward primer MusF1 (GGTACCCGGAGTGGGTTC) and reverse primer MusR (CGAGCTGTCCACATTAGAAA) for mouse cDNA and forward primers HsaF1 (TCCTGAATTCCTGTGGCTGT), HsaF2 (ATGGAGAAGACAGGCTTTCG), and HsaF3 (ATGAGGAGCTTGAGGAAGCA) and reverse primer HsaR1 (ATCTTGACCCCTGGAGCTTT) for human cDNA. The PCR reaction began with a 2-min hot start, followed by a six-cycle touchdown: 95°C for 30 s, 66°C for 30 s, and 72°C for 1 min. The annealing temperature decreased 1°C in each of the six cycles. The reaction was then carried out an additional 30 cycles, with each cycle consisting of the following: 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. The reaction was then held at 72°C for 7 min. The PCR products were separated on a 2% agarose gel and visualized by ethidium bromide.
Animals used as the source of mouse RNA were housed and handled according to NIH Guidelines for the Care and Use of Laboratory Animals under UCSF Protocols AN076327 and AN81551.
Quantitative reverse-transcriptase PCR
Specific primer and probe sets were designed through Assays-by-Design (Applied Biosystems, Foster City, CA) for the full-length and alternative (internal promoter) human INPP5B transcripts. For the full-length transcript, primers were as follows: forward primer was HsaF1, reverse primer was HsaR2, and the probe was ACCTCCGCCAATTGT, which spans the GC-AG splice site used in the human full-length transcript and is absent if the GT splice site is used. For qPCR of the alternative transcript using the internal promoter, primers were as follows: forward primer was HsaF3, reverse primer was GAACCACACCTGCAGTGTTG, which spanned the GT-AG splice site used in the alternative transcript and is absent if the GC splice site is used, and the probe was AAATGTCTGCCGCCGCCG, which is present in exon 1 of the alternative transcript but is part of intron 7 and therefore absent from the mature mRNA of the full-length transcript. Relative expression of the two INPP5B transcripts was obtained versus human PGK1 (Hs99999906_m1).
Each assay was performed in triplicate following the ABI protocol. Ten nanograms of input RNA was used per assay. For all assays, the PCR was first held at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The critical threshold (C T) was identified in the linear phase of the amplification. The C T was then used for relative quantitation analysis. Relative quantitation was done according to the Comparative C T Method described in Applied Biosystems User Bulletin #2 for Relative Quantitation of Gene Expression. Prior to analysis, each assay was first compared to the PGK1 control to demonstrate equal efficiency of target amplification described in User Bulletin #2. The absolute value of the slope of log input amount versus DCT was less than 0.1 for each assay, indicating a similar efficiency of approximately 2 between assays.
In silico promoter prediction
PROSCAN ver. 1.7 (Prestridge 1995) was used to scan for possible transcription factor sites. Using a cutoff score of 53, the program has an approximately 70% sensitivity for recognizing primate promoter sequences and a false-positive rate of about 0.0007% per base.
Luciferase assay
To test for internal promoter function, a genomic fragment encompassing most of mouse Inpp5b exon 7 and extending 42 bp into intron 7 (Chr4 +strand 124,428,509–124,428,978; NCBI37/mm9) and a similar fragment encompassing most of human INPP5B exon 7 and extending into intron 7 (Chr1 −strand 38,397,369–38,397,703; CRCh37/Hg19) were isolated by PCR and cloned separately into the promoterless basic pGL4.10 reporter vector (Promega, Madison, WI). pGL4.13 containing the SV40 promoter and pGL4.10 without any cloned material served as a positive control and a promoterless negative control, respectively. Cotransfection with a fixed amount of pGL4.73 expressing renilla from an SV40 promoter served as a control for transfection efficiency. Plasmids were transfected in triplicate per experiment and the ratio of luciferase to renilla activity was determined as described previously (Sotiriou et al. 2009).
Results
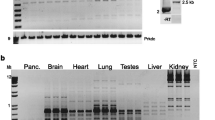
Previously published Northern blot analyses using the full-length murine Inpp5b cDNA (GenBank NM_008385) as probe revealed that the mouse gene generates two readily detectable transcripts in most tissues examined (Janne et al. 1998; Matzaris et al. 1998). These differ in size by approximately 800 bp and are present in approximately equal amounts in many tissues, including lung, liver, and kidney, but differ in amount relative to each other in other tissues such as brain, spleen, or testis. A review of the reference mRNA sequences of mouse Inpp5b in NCBI Entrez reveal two isoforms: a larger 3.826-kb transcript, consisting of 24 exons (GenBank entry NM_008385) that encodes a protein of 993 amino acids (GenBank entry NP_032411), and a shorter transcript of 2.964 kb (GenBank entry AK004722) that is identical to the larger transcript in exons 8 through 24 but lacks exons 1–6 and the first two thirds of exon 7. The difference between these transcripts was confirmed experimentally by Northern blotting: a cDNA probe containing only the first six exons of Inpp5b detects only the larger approximately 3.8-kb transcript (Fig. 1). The simplest explanation for these findings is that the shorter isoform is derived from a transcriptional start site at an alternative internal promoter located within exon 7.
Northern blot analysis of mouse RNA using the first six exons of Inpp5b as a probe. The arrowhead marks the ~4 kb expected for the full-length transcript, while the arrow marked with an asterisk is where the shorter transcript would ordinarily be if a full-length probe had been used. Lane 1 heart; lane 2 brain; lane 3 liver; lane 4 spleen; lane 5 kidney; lane 6 total embryo; lane 7 lung; lane 8 thymus; lane 9 testes; lane 10 ovary
In another previously published Northern blot, humans, in contrast, showed only a single approximately 4.5-kb INPP5B transcript, including in tissues relevant to Lowe syndrome, such as brain and kidney (Janne et al. 1998). This transcript corresponds to GenBank entry NM_005540, which is 4.469 kb in length and encodes a 913-residue human Inpp5b enzyme (GenBank entry NP_005531). A comparison of the amino acid sequence of the human enzyme (NP_005531) and the larger of the two mouse enzymes (NP_032411) shows a high degree of similarity between segments encoded by exons 1–6 (90% identical, 93% conserved) and exons 9 through to the end of the gene (88% identical and 94% conserved). However, there are significant differences in the protein segments encoded by exons 8 and 7. The amino acid sequences of the segments encoded by exon 8 are the same size in both species but only 46% identical and 57% conserved. Even more striking is the difference in the cDNA and protein representing exon 7. The segment of the protein encoded by human exon 7 is 47 amino acids long while that from mouse contains 127 amino acids, a difference of 80 amino acids (Fig. 2a). Examination of genomic DNA and transcripts shows that exon 7 in mouse is 381 bp long while human exon 7 is only 141 bp (Fig. 2b). There is nucleic acid homology not only between the first 141 bases of mouse exon 7 and all 141 bases of human exon 7, but also between the remaining 240 bp of mouse exon 7 and the first 240 bp of human intron 7. The expected canonical GT 5′ splice site at the beginning of mouse intron 7 is conserved in human genomic DNA, at the +240 position inside human intron 7, while a noncanonical GC 5′ splice donor site that marks the end of human exon 7 is conserved in the mouse but located within exon 7 at the orthologous location 141 bases from the beginning of the exon. We infer from these data that human exon 7 results from utilization of the internal GC 5′ site, while mouse exon 7 is defined by a GT 5′ splice site 240 bases further downstream.
a Alignment of segments of human and mouse Inpp5b enzyme encoded by exons 7 and 8. The segments corresponding to exons 7 and 8 are shaded as indicated. b Alignment of human and mouse exons 7, the portion of human intron 7 homologous to mouse exon 7, and exon 8. Segments are shaded with the same patterns used in a, indicating the various portions of exon 7 and exon 8. The location of primers discussed in the text is shown. The bent arrow indicates the approximate location of the transcriptional start of the transcripts generated from the internal promoter in mouse exon 7 and the analogous region in human intron 7
We confirmed the difference in splice donor site usage in the two species by reverse transcriptase PCR (RT-PCR). For the mouse, we chose a forward primer (MusF1) located at the beginning of exon 7 and a reverse primer (MusR) in exon 8 (Fig. 2b). For the human, we used three human-specific forward primers (HsaF1, HsaF2, and HsaF3) and one reverse (HsaR1) PCR primer. Primer HsaF1 is upstream of the GC splice site in exon 7 (Fig. 2b), HsaF2 is situated in human intron 7 just downstream of the GC 5′ splice site, and HsaF3 is located further down in human intron 7 in the region just upstream of the GT dinucleotide that is homologous to the 5′ splice site used by the mouse. HsaR1 is in exon 8.
RT-PCR of mouse brain and kidney RNA using MusF1 and MusR generated only the 297-bp fragment expected if the GT splice site were used and never the much smaller fragment corresponding to splicing at the GC site (Fig. 3a). In contrast, RT-PCR of human brain or kidney RNA with the human HsaF1 and HsaR1 primers produced only a 186-bp fragment, as expected if splicing occurred exclusively at the GC site, and not the larger 426-bp product that would be predicted if splicing in humans occurred at the GT 5′ splice site within intron 7 (Fig. 3b, c). To confirm this result, we used RT-PCR with a second forward primer (HsaF2) located immediately downstream of the GC splice site. RT-PCR with HsaF2 and HsaR1 failed to generate any product in human brain or kidney RNA (Fig. 3b, c), thereby demonstrating that the GC 5′ splice site and not the downstream GT site, which corresponds to the site used exclusively by the mouse, is always used when exon 7 is transcribed in these tissues. The failure to detect a fragment with HsaF2-HsaR1 was not the result of nonfunctional primers since a product of the correct size was seen using as template an artificial cDNA construct (labeled artificial ex7 + 8 in Fig. 3) containing a segment of genomic DNA that included all of human exon 7 plus the first 240 bp of intron 7 ligated to a segment of genomic DNA containing exon 8.
a Semiquantitative reverse transcriptase PCR of murine brain (B) and kidney (K) RNA using primers MusF1 and MusR for 25, 30, and 35 cycles. A 50-bp DNA ladder is shown on the left with the 200- and 350-bp fragments labeled for orientation purposes. Only a 297-bp fragment corresponding to splicing at the GT splice site was seen with RT-PCR of mouse Inpp5b using MusF1 and MusR primers. b Reverse transcriptase PCR of human brain RNA using primers HsaF1, HsaF2, and HsaF3 and reverse primer HsaR1. A 50-bp DNA ladder is shown on the left with the 200- and 350-bp fragments labeled for orientation purposes. A negative control in which the reverse transcriptase was omitted (-RT) is shown. A 192-bp fragment corresponding to splicing at the GC splice site was seen with HsaF1 and HsaR1 primers, while a 146-bp fragment was seen with HsaF3 and HsaR1. PCR of an artificial DNA construct, ex7 + ex8, which consists of a segment of genomic DNA containing human exon 7 and the first 240 bp of intron 7 ligated to a segment of genomic DNA containing exon 8, is shown as a positive control for all three forward human primers, particularly HsaF2. c Same as B except kidney RNA was used
Given the absence of any RT-PCR product from human RNA using primers HsaF2 and HsaR1 (Fig. 2b), we were surprised to find that a third pair of primers, HsaF3-HsaR1, did give an RT-PCR product of 145 bp in human brain and kidney RNA (Fig. 3b, c). The product was sequenced and shown to be cDNA sequence from the distal portion of the first 240 bp of human intron 7, showing homology to mouse exon 7 joined to exon 8. This product was therefore the product of splicing at the GT dinucleotide homologous to the mouse 5′ splice site. We reasoned that this RT-PCR product from human intron 7 could not be due to a full-length human transcript in human that mimicked mouse splicing because the HsaF1-HsaR1 RT-PCR product was only 194 bp, indicating that only the GC 5′ splice site was being used, and no RT-PCR product was seen using primer pair HsaF2-HsaR1. We therefore concluded that, like the smaller transcript in mouse that was detectable by Northern blot analysis, a smaller transcript was also made in humans that had not been previously detected by Northern blot analysis. This transcript likely initiated at an internal promoter located within the first 240 bp of intron 7 and then utilized the GT dinucleotide homologous to the mouse 5′ splice site to splice onto exon 8. Indeed, an examination of human INPP5B transcripts in GenBank reveals a 757-bp isoform (AK022846) as well as ten expressed sequence tags (Supplementary Table 1) in which the 5′ ends of the transcripts contain sequence from intron 7 spliced onto exon 8. A diagram summarizing all the RT-PCR results for exons 7 and 8 in INPP5B and Inpp5b is shown in Fig. 4.
The region in mouse exon 7 and human intron 7 containing the putative internal promoter is GC rich and contains numerous transcription factor binding sites. A promoter scan of the forward strand (Proscan 1.7) gave a promoter score of 98.89 in humans and 74.73 in mice, far above the cutoff score of 53 in both species. We therefore decided to test this segment of DNA for promoter function in a transient transfection assay with a luciferase reporter. Promoter activity of a segment from either the 3′ end of mouse exon 7 or the homologous human intron 7 sequence was assessed in a transient luciferase assay. Segments from both species had promoter activity equal to or exceeding the SV40 promoter in the control pGL4 luciferase vector (Fig. 5). The approximate location of the transcriptional start based on GenBank entries AK022846 for human and AK004722 in mouse is shown in Fig. 3, but the diagram is only an approximation since actual transcript mapping of the start of transcription has not been done.
Because the human INPP5B alternative transcript made from the internal promoter was seen only by a sensitive RT-PCR method and not on Northern blot analysis, we used quantitative RT-PCR (qRT-PCR) to measure more accurately the amount of alternative transcript made from the internal promoter relative to the full-length transcript in humans. Real-time qRT-PCR assays using primers and probes (TaqMan®, Applied Biosystems) specific for the full-length human transcript and the shorter alternative transcript showed that the alternative transcript was a minor transcript in most tissues (Fig. 6). Except for testis and spleen, in which the shorter transcript was 50 and 20% of total INPP5B, respectively, it constituted only about 5–10% of total INPP5B mRNA in lung, liver, and retina, and about 3% or less of total INPP5B in such tissues as brain and kidney that are relevant to Lowe syndrome. This explains why the shorter alternative transcript was difficult to visualize on Northern blot analysis of human mRNA. The low abundance of this transcript contrasts with what is seen in mouse, where there is sufficient transcript originating from the internal promoter to allow the smaller transcript to be readily detected on Northern blot in a number of tissues.
Discussion
In seeking to explain the phenotypic disparity between mice lacking Ocrl, which are normal, and humans lacking OCRL, who have Lowe syndrome, we eliminated interspecies differences in the OCRL/Ocrl genes themselves. First, the human and mouse OCRL/Ocrl orthologs are 91% identical and 95% similar at the amino acid level. Both are expressed highly in nearly all tissues except lymphocytes in both species (Janne et al. 1998; Olivos-Glander et al. 1995). The only alternative splicing is conserved in the two species, resulting in inclusion or exclusion of a highly homologous 24-base exon (NM_000276.3 and NM_0001587.3, respectively, in human, NM_177215.3 and numerous ESTs, respectively, in mouse) located at the same position in the mRNA. There is also a highly unusual splice acceptor site, an AT, at the 3′ end of intron 2 in both human and mouse in OCRL and Ocrl that serves as acceptor for a canonical GT 5′ splice donor. Otherwise, OCRL and Ocrl splicing is constitutive, uses canonical sites, and generates exons of identical size in both species.
In contrast, the autosomal paralogs, mouse Inpp5b and human INPP5B, which are the closest paralogs to Ocrl in the genome with many structural motifs in common with it, differ in a number of ways that might explain why the two species compensate for an Ocrl enzyme deficiency differently. INPP5B and Inpp5b are the only PtdIns 5-phosphatases to have a Rho-GAP domain along with the shared inositol phosphate 5-phosphatase domain. The rho-GAP domain is noncatalytically active but has affinity for the Rac (Faucherre et al. 2003) and ARF (Lichter-Konecki et al. 2006) small G proteins. The paralogs also share a pleckstrin-homology domain (Mao et al. 2009) and an ASH (ASPM, SPD-2, Hydin) domain (Ponting 2006). The ASH and Rho-GAP domains together interact with APPL1, a Rab5 effector protein (Erdmann et al. 2007; McCrea et al. 2008), and the endosomal proteins Ses1 and Ses2 (Swan et al. 2010). Ocrl, however, has a clathrin-binding domain (Choudhury et al. 2005, 2009) that is absent from Inpp5b (Erdmann et al. 2007). Inpp5b has similar intracellular localization to Ocrl (Erdmann et al. 2007) as well as similar, but not identical, substrate specificity for PtdIns(4,5)P2 (Janne et al. 1998; Schmid et al. 2004). Finally, although cellular assays ex vivo suggest that Inpp5b is not capable of complementing all of Ocrl’s functionality (Coon et al. 2009; Williams et al. 2007), genetic analysis indicates that Ocrl and Inpp5b have overlapping function in vivo (Bernard and Nussbaum 2010; Janne et al. 1998).
We did find differences in transcription, splicing, and sequence between INPP5B and Inpp5b: (1) The full-length human Inpp5b enzyme lacks 80 amino acids because a GC splice site is used that causes 240 bp of sequence that is evolutionarily conserved with mouse exon 7 to be treated as intron and spliced out; (2) relative to the respective full-length transcripts, there is a reduced amount of a shorter alternative transcript originating from an internal promoter in humans compared to mouse; and (3) exon 8 is more divergent than the rest of the enzyme in the two species.
The reasons for the difference in 5′ splice site utilization between mouse Inpp5b and human INPP5B are unknown. Among mammalian splice sites, 98.71% contain canonical GT-AG junctions while only 0.56% has a noncanonical GC-AG splice-site pair (Burset et al. 2001). A GC 5′ splice site is preceded by a G, as it is in the human, 97.6% of the time, and by an A, as it is in C57Bl/6 and most inbred mouse strains, only 1.6% of the time. Of interest, rat (R. norvegicus) and bovine (B. taurus) Inpp5b also splice predominantly at the GC splice site rather than the GT site the mouse uses and both of these mammals have a G immediately preceding the GC splice donor. However, this difference is not sufficient to explain why mouse does not use the internal GC 5′ donor because sequencing of 12 inbred strains of mice revealed that AKR/J mice and their descendants have a G preceding the internal GC splice site and yet still do not use this splice site in liver, brain, and kidney mRNA (data not shown). Thus, the exclusive use of the downstream GT splice site by mouse seems unique to the mouse and not present in human, bovine, and rat species.
Conversely, at the GT 5′ splice site utilized by mice, both humans and mice have the typical CAG immediately preceding the 5′ splice site, both have the canonical GT at positions +1 and +2, both have the highly preferred (82%) G at the +5 position, and neither has the preferred A or G at +3 or the preferred A at +4 (Cartegni et al. 2002). The Analyzer Splice Tool, which calculates scores for 5′ and 3′ splice sites (Shapiro and Senapathy 1987), gives very comparable scores for the mouse GT 5′ splice site and the analogous human site of 79.78 and 73.91, respectively. The human GT 5′ splice site can clearly be functional since it is used to generate the shorter alternative transcript in humans. Conversely, the human GC 5′ splice site and the analogous mouse GC dinucleotide from the AKR/J mice have comparable scores of 69.73 and 68.92, although the score when the GC is preceded by an A, as it is in most inbred mouse strains, is much less. Thus, the immediate contexts around the GT and GC 5′ splice sites are not significantly different between the two species and do not explain the difference in splice-site usage. Furthermore, the 3′ splice sites at the end of intron 7 also have comparable scores of 83.32 and 88.37, respectively, in human and mouse. Finally, intron sizes are large in both species (25,649 in mouse and 40,459 in human) and therefore provide no restraint on the utilization of the GT splice site in humans. Despite the lack of clear sequence differences that would explain the preference for the GC versus the GT 5′ splice site in human INPP5B, the phenomenon is a cis and not a trans effect since human INPP5B integrated into the mouse genome on a bacterial artificial chromosome in a transgenic mouse (R. L. Nussbaum, unpublished) shows the same overwhelming preference for the GC splice site (Supplementary Fig. 1).
The biochemical and cellular functional consequences of the interspecies differences between the human and mouse orthologs of Inpp5b remain to be elucidated. Nonetheless, they represent a significant interspecies difference in a potential modifying paralog for Ocrl that might explain why Inpp5b and INPP5B differ in their ability to compensate for loss of Ocrl function. These results set the stage for investigating which of the differences are responsible for the disparate phenotype in Ocrl-deficient humans and mice, which may, in turn, provide additional insights into the pathogenesis of Lowe syndrome in humans.
References
Attree O, Olivos IM, Okabe I, Bailey LC, Nelson DL et al (1992) The Lowe oculocerebrorenal syndrome gene encodes a novel protein highly homologous to inositol polyphosphate-5-phosphatase. Nature 358:239–242
Bernard DJ, Nussbaum RL (2010) X-inactivation analysis of embryonic lethality in Ocrl wt/− ;Inpp5b −/− mice. Mamm Genome 21:186–194
Bockenhauer D, Bokenkamp A, van’t Hoff W, Levtchenko E, van Kist-Holthe JE et al (2008) Renal phenotype in Lowe Syndrome: a selective proximal tubular dysfunction. Clin J Am Soc Nephrol 3:1430–1436
Burset M, Seledtsov IA, Solovyev VV (2001) SpliceDB: database of canonical and non-canonical mammalian splice sites. Nucleic Acids Res 29:255–259
Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298
Charnas L, Bernardini I, Rader D, Hoeg J, Gahl WA (1991) Clinical and laboratory findings in the oculocerebrorenal syndrome of Lowe, with special reference to growth and renal function. N Engl J Med 324:1318–1325
Choudhury R, Diao A, Zhang F, Eisenberg E, Saint-Pol A et al (2005) Lowe syndrome protein OCRL1 interacts with clathrin and regulates protein trafficking between endosomes and the trans-Golgi network. Mol Biol Cell 16:3467–3479
Choudhury R, Noakes CJ, McKenzie E, Kox C, Lowe M (2009) Differential clathrin binding and subcellular localization of OCRL1 splice isoforms. J Biol Chem 284:9965–9973
Coon BG, Mukherjee D, Hanna CB, Riese DJ 2nd, Lowe M et al (2009) Lowe syndrome patient fibroblasts display Ocrl1-specific cell migration defects that cannot be rescued by the homologous Inpp5b phosphatase. Hum Mol Genet 18:4478–4491
Erdmann KS, Mao Y, McCrea HJ, Zoncu R, Lee S et al (2007) A role of the Lowe syndrome protein OCRL in early steps of the endocytic pathway. Dev Cell 13:377–390
Faucherre A, Desbois P, Satre V, Lunardi J, Dorseuil O et al (2003) Lowe syndrome protein OCRL1 interacts with Rac GTPase in the trans-Golgi network. Hum Mol Genet 12:2449–2456
Janne PA, Suchy SF, Bernard DJ, McDonald M, Crawley J et al (1998) Functional overlap between murine Inpp5b and Ocrl1 may explain why deficiency of the murine ortholog for OCRL1 does not cause Lowe syndrome in mice. J Clin Invest 101:2042–2053
Kenworthy L, Charnas L (1995) Evidence for a discrete behavioral phenotype in the Oculocerebrorenal Syndrome of Lowe. Am J Med Genet 59:283–290
Kenworthy L, Park T, Charnas LR (1993) Cognitive and behavioral profile of the Oculocerebrorenal Syndrome of Lowe. Am J Med Genet 46:297–303
Leahey AM, Charnas LR, Nussbaum RL (1993) Nonsense mutations in the OCRL-1 gene in patients with the Oculocerebrorenal Syndrome of Lowe. Hum Mol Genet 4:461–463
Lichter-Konecki U, Farber LW, Cronin JS, Suchy SF, Nussbaum RL (2006) The effect of missense mutations in the RhoGAP-homology domain on ocrl1 function. Mol Genet Metab 89:121–128
Lin T, Orrison BM, Suchy SF, Lewis RA, Nussbaum RL (1998) Mutations are not uniformly distributed throughout the OCRL1 gene in Lowe Syndrome patients. Mol Genet Metab 64:58–61
Mao Y, Balkin DM, Zoncu R, Erdmann KS, Tomasini L et al (2009) A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J 28:1831–1842
Matzaris M, O’Malley CJ, Badger A, Speed CJ, Bird PI et al (1998) Distinct membrane and cytosolic forms of inositol polyphosphate 5-phosphatase II. J Biol Chem 273:8256–8267
McCrea HJ, Paradise S, Tomasini L, Addis M, Melis MA et al (2008) All known patient mutations in the ASH-RhoGAP domains of OCRL affect targeting and APPL1 binding. Biochem Biophys Res Commun 369:493–499
Monnier N, Satre V, Lerouge E, Berthoin F, Lunardi J (2000) OCRL1 mutation analysis in French Lowe syndrome patients: implications for molecular diagnosis strategy and genetic counseling. Hum Mutat 16:157–165
Olivos-Glander IM, Janne PA, Nussbaum RL (1995) The Oculocerebrorenal Syndrome gene product is a 105-kDa protein localized to the Golgi complex. Am J Hum Genet 57:817–823
Ponting CP (2006) A novel domain suggests a ciliary function for ASPM, a brain size determining gene. Bioinformatics 22:1031–1035
Prestridge DS (1995) Predicting Pol II promoter sequences using transcription factor binding sites. J Mol Biol 249:923–932
Schmid AC, Wise HM, Mitchell CA, Nussbaum R, Woscholski R (2004) Type II phosphoinositide 5-phosphatases have unique sensitivities towards fatty acid composition and head group phosphorylation. FEBS Lett 576:9–13
Shapiro MB, Senapathy P (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res 15:7155–7174
Sotiriou S, Gibney G, Baxevanis AD, Nussbaum RL (2009) A single nucleotide polymorphism in the 3′UTR of the SNCA gene encoding alpha-synuclein is a new potential susceptibility locus for Parkinson disease. Neurosci Lett 461:196–201
Suchy SF, Nussbaum RL (2009) Oculocerebrorenal Syndrome of Lowe. In: Valle D, Vogelstein B, Kinzler K, Antonarakis S, Ballabio A (eds) The online metabolic and molecular bases of inherited disease. MacMillan, New York
Suchy SF, Olivos-Glander IM, Nussbaum RL (1995) Lowe Syndrome, a deficiency of a phosphatidylinositol 4,5-bisphosphate 5-phosphatase in the Golgi apparatus. Hum Mol Genet 4:2245–2250
Swan LE, Tomasini L, Pirruccello M, Lunardi J, De Camilli P (2010) Two closely related endocytic proteins that share a common OCRL-binding motif with APPL1. Proc Natl Acad Sci USA 107:3511–3516
Williams C, Choudhury R, McKenzie E, Lowe M (2007) Targeting of the type II inositol polyphosphate 5-phosphatase INPP5B to the early secretory pathway. J Cell Sci 120:3941–3951
Zhang X, Jefferson AB, Auethavekiat V, Majerus PW (1995) The protein deficient in Lowe syndrome is a phosphatidylinositol-4,5-bisphosphate 5-phosphatase. Proc Natl Acad Sci USA 92:4853–4856
Acknowledgments
This study was generously supported by The Lowe Syndrome Trust, UK (SPB and RLN), intramural NHGRI/NIH (LWF), and institutional support from the Department of Medicine and the Institute for Human Genetics, UCSF (SPB, AH, RLN). We are grateful to Dr. Yien-Ming Kuo for help in preparing the manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
335_2010_9281_MOESM2_ESM.tif
Supplementary Figure 1 RT PCR of RNA from human fibroblasts (HuFib), from mouse spleen and mouse brain from a non-transgenic mouse, and from the same two tissues from two independent transgenic mouse lines expressing human INPP5B from a BAC insertion. HsaF1 and HsaR1 were used for the PCR. Arrowhead marks the size (186 bp) of the human product expected if the usual human GC splice site is used, while the arrow with asterisk is the location of where the 297 bp product would be if the GT splice site had been used (TIFF 1026 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Bothwell, S.P., Farber, L.W., Hoagland, A. et al. Species-specific difference in expression and splice-site choice in Inpp5b, an inositol polyphosphate 5-phosphatase paralogous to the enzyme deficient in Lowe Syndrome. Mamm Genome 21, 458–466 (2010). https://doi.org/10.1007/s00335-010-9281-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-010-9281-7