Abstract
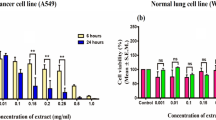
The aim of the present research was to study the anticancer effects of Aspergillus niger (A.niger) RNase. We found that RNase (A.niger RNase) significantly and dose dependently inhibited invasiveness of breast cancer cell line MDA MB 231 by 55 % (P < 0.01) at 1 μM concentration. At a concentration of 2 μM, the anti invasive effect of the enzyme increased to 90 % (P < 0.002). Keeping the aim to determine molecular level interactions (molecular simulations and protein docking) of human actin with A.niger RNase we extended our work in-vitro to in-silico studies. To gain better relaxation and accurate arrangement of atoms, refinement was done on the human actin and A.niger RNase by energy minimization (EM) and molecular dynamics (MD) simulations using 43A2 force field of Gromacs96 implemented in the Gromacs 4.0.5 package, finally the interaction energies were calculated by protein-protein docking using the HEX. These in vitro and in-silico structural studies prove the effective inhibition of actin activity by A.niger RNase in neoplastic cells and thereby provide new insights for the development of novel anti cancer drugs.








Similar content being viewed by others
References
Vasandani VM, Wu YN, Mikulski SM, Youle RJ, Sung C (1996) Molecular determinants in the plasma clearance and tissue distribution of ribonucleases of the ribonuclease A superfamily. Cancer Res 56:4180–4186
Darzynkiewicz Z, Carter SP, Mikulski SM, Ardelt WJ, Shogen K (1988) Cytostatic and cytotoxic effects of Pannon (P-30 Protein), a novel anticancer agent. Cell Tissue Kinet 21:169–182
Mikulski SM, Viera A, Darzynkiewicz Z, Shogen K (1992) Synergism between a novel amphibian oocyte ribonuclease and lovastatin in inducing cytostatic and cytotoxic effects in human lung and pancreatic carcinoma cell lines. Br J Cancer 66:304–310
Ryback SM, Pearson JW, Fogler WF, Volker K, Spence SE et al (1996) Enhancement of vincristine cytotoxicity in drug-resistant cells by simultaneous treatment with onconase, an antitumor ribonuclease. J Natl Cancer Inst 88:747–753
Deptala A, Halicka HD, Ardelt W, Mikulski SM, Shogen K et al (1998) Potentiation of tumor necrosis factor induced apoptosis by onconase. Int J Oncol 13:11–16
Matousek J, Soucek J, Slavik T, Tomanek M, Lee JE et al (2003) Comprehensive comparison of the cytotoxic activities of onconase and bovine seminal ribonuclease. Comput Biochem Physiol C 136:343–356
Mikulski SM, Costanzi JL, Vogelzang NJ, McCachren S, Taub RN et al (2002) Phase II trial of a single weekly intravenous dose of ranpirnase in patients with unresectable malignant mesothelioma. J Clin Oncol 20:274–281
Schwartz B, Shoseyov O, Melnikova VO, McCarty M, Leslie M et al (2007) ACTIBIND, a T2 RNase, competes with angiogenin and inhibits human melanoma growth, angiogenesis, and metastasis. Cancer Res 67:5258–5266
Zhang Y, Li W, Su Z (2001) Trends in the pharmaceutical research of ribonucleases and their therapeutic uses. J Biomed Eng 18:456–460
Makarov AA, Ilinkaya O (2003) Cytotoxic ribonucleases: molecular weapons and their targets. FEBS Lett 540:15–20
Makarov AA, Kolchinski A, Ilinskaya ON (2008) Binase and other microbial RNases as potential anticancer agents. BioEssays 30:789–790
Saxena SK, Gravell M, Wu Y (1996) Inhibition of HIV-1 production and selective degradation of viral RNA by an amphibian ribonuclease. J Biol Chem 271:20783–20788
Skvortsova MA, Bocharov AL, Yakovley GI (2002) Novel extracellular ribonuclease from Bacillus intermedius-binase II: purification and some properties of the enzyme. Biochem Mosc 67:802–806
Guan GP, Wang HX, Ng TB (2007) A novel ribonuclease with antiproliferative activity from fresh fruiting bodies of the edible mushroom Hypsizigus marmoreus. Biochim Biophys Acta 1770:1593–1597
Lam SK, Ng TB (2001) Isolation of a novel thermolabile heterodimeric ribonuclease with antifungal and antiproliferative activities from roots of the Sanchi Ginseng Panax otoginseng. Biochem Biophys Res Commun 285:419–423
Lindblom M, Morgen H (1974) Enzymatic RNA reduction in disintegrated cell Saccharomyces cerevisia. Biotechnol Bioeng 16:1123–1133
Gundampati RK, Rajasekhar C, Moni K, Anurag S, Pratyush DD et al (2011) Protein-protein docking on molecular models of Aspergillus niger RNase and human actin: novel target for anticancer therapeutics. J Mol Model 18:653–662
Gundampati RK, Sharma A, Kumar M, Debnath M (2011) Extracellular poly (A) specific ribonuclease from Aspergillus niger ATCC 26550: purification, biochemical, and spectroscopic studies. Process Biochem 46:135–141
Patricia CP, Rafael AC, Fernanda C, Walter FA (2009) Molecular modeling dynamics simulation of human cyclin-dependent kinase 3 complexed with inhibitors. Comput Biol Med 39:130–140
Arfken G (1985) The method of steepest descents.n§7.4. In: Mathematical methods for physicists, 3rd edn. Academic, Orlando, pp 428–436
Berendsen HJC, Grigera JR, Straatsma TP (1987) The missing term in effective pair potentials. J Phys Chem 91:6269–6271
Chowdhuri S, Tan ML, Ichiye TJ (2006) Dynamical properties of the soft sticky dipole–quadrupole–octupole water model: a molecular dynamics study. J Chem Phys 125:14451–14453
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Macindoe G, Mavridis L, Venkatraman V, Devignes MD, Ritchie DW (2010) Hex server: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res 38:445–449. doi:10.1093/nar/gkq311
Humphrey W, Dalke A, Schulten K (1996) VMD-visual molecular dynamics. J Mol Graph 14:33–38
Lichtarge O, Bourne HR, Cohen FE (1996) An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol 257:342–358
Jones S, Thronton JM (1997) Analysis of protein-protein interaction sites using surface patches. J Mol Biol 272:121–132
Jones S, Thromton JM (1997) Prediction of protein protein interaction sites using patches analysis. J Mol Biol 272:133–143
Neuvirth H, Raz R, Schreiber G (2004) Promote: a structure based prediction program to indentify the location of protein-protein binding sites. J Mol Biol 338:181–199
Bradford J, Westhead D (2005) Improved prediction of protein-protein binding sites using a support vector machines approach. Bioinformatics 211:487–1494
Zhou H, Shan Y (2001) Prediction of protein interaction sites from sequence profile and residue neighbor list. Proteins 44:336–343
Espadaler J, Romero-Isart O, Jackson RM, Oliva B (2005) Prediction of protein-protein interactions using distant conservation of sequence patterns and structure relationships. Bioinformatics 21:3360–3368
Aytuna AS, Gursoy A, Keskin O (2005) Prediction of protein-protein interactions by combining structure and sequence conservation in protein interfaces. Bioinformatics 2:2850–2855
Acknowledgments
GRK, SS and NG wants to thanks University Grants Commission, Government of India for financial support and the School of Biochemical Engineering, Institute of Technology, BHU, Varanasi, India. KS wishes to thank Council of Scientific & Industrial Research, Government of India for providing financial assistance. SL wants to thank Indian Council of Medical Research (ICMR),New Delhi, Government of India for financial support and Tumour Biology Laboratory, National Institute of Pathology, Safdarjung Hospital Campus, New Delhi-110029 and RSC, CM gratefully acknowledges University Grant Commission (UGC), New Delhi (F.No.33-222/2007 (SR) 13-3-08) for financial support. The encouragement given by Sri Krishnadevaraya University, AP, India to our research work is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Gundampati Ravi Kumar, Rajasekhar Chikati and Santhi Latha Pandrangi contributed equally to this work.
Rights and permissions
About this article
Cite this article
Kumar, G.R., Chikati, R., Pandrangi, S.L. et al. Molecular docking and dynamics simulations of A.niger RNase from Aspergillus niger ATCC26550: for potential prevention of human cancer. J Mol Model 19, 613–621 (2013). https://doi.org/10.1007/s00894-012-1587-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1587-9