Abstract
Kawasaki disease (KD) is an acute systemic vasculitis syndrome that primarily affects infants and young children. The cause of KD is largely unknown, but its higher incidence in the Asian population and increased risk in patients’ families suggests the existence of underlying genetic factors. To determine the loci of a susceptibility gene for KD, a genomewide linkage analysis with affected sib pairs was performed on 78 family samples collected from all over Japan. Multipoint linkage analysis using MAPMAKER/SIBS 2.0 identified evidence of linkage on 12q24 [maximum lod score (MLS)=2.69]. Possible linkage (MLS > 1.0) was also found on 4q35, 5q34, 6q27, 7p15, 8q24, 18q23, 19q13, Xp22, and Xq27. This is the first large-scale study of the genetic susceptibility to KD, and our results, combined with the accumulated knowledge of the human genome, could greatly promote research on identification of the molecular pathogenesis of KD.
Similar content being viewed by others
Introduction
Kawasaki disease (KD; MIM300530), also known as mucocutaneous lymph node syndrome (MCLS), was first described in 1967 by the Japanese pediatrician Tomisaku Kawasaki. It is a self-limiting vasculitis syndrome that affects systemic small and medium-sized blood vessels. The symptoms include prolonged fever, nonsuppurative cervical lymphadenitis, and changes in the skin and mucous membranes, such as rash; edema; conjunctival injection; erythema of the oral cavity, lips, and palms; and desquamation of the fingertips (Kawasaki 1967; Burns 2002). Although the incidence has been dramatically lowered by introduction of high-dose immunoglobulin therapy, 15–20% of untreated patients and patients resistant to the therapy develop serious, sometimes life-threatening, cardiac sequelae associated with coronary artery aneurysms (Kato et al. 1975). In Japan, approximately 0.08% of patients die in the acute phase of the disease, mostly due to cardiac complications (Yanagawa et al. 1998). Furthermore, patients with cardiac sequelae have an increased risk of ischemic heart disease that may lead to myocardial infarction and sudden death (Kato et al. 1996). Currently, KD is the most common cause of acquired heart disease in childhood in the developed countries. Despite intensive study over more than 35 years, the etiology of the disease has not been clarified. In Japan, 18 nationwide epidemiologic surveys of KD have been carried out biennially from 1970 to the present. These surveys have revealed several important features, such as: (1) unidentified infectious agent(s) appear to play some role, (2) siblings and offspring of KD patients are at higher risk of the disease (Fujita et al. 1989; Uehara et al. 2003), and (3) male predominance of its occurrence (male:female ratio 1.4) and cardiac complications (Yanagawa et al. 1998). These features strongly indicate the existence of genetic factors that determine the susceptibility and severity of KD, in addition to any environmental factors. A possible mechanism of KD onset would be a genetically determined exaggerated immune response triggered by some unknown infection. Thus, identification of genetic factors would greatly facilitate understanding of the disease etiology and pathophysiology. We conducted a nonparametric genome-wide linkage analysis on 78 Japanese affected sib pairs (trios), and several candidate gene loci have been identified.
Materials and methods
Family samples
Siblings affected with KD were recruited nationwide for this study. All patients were diagnosed by pediatricians according to the criteria established by the Japan Kawasaki Disease Research Committee (http://www.kawasaki-disease.org/diagnostic/index.html). Genomic DNA was extracted from peripheral blood leukocytes or Epstein-Barr-virus-transformed lymphoblastoid cell lines. The Ethical Committee of the Institute of Medical Science, the University of Tokyo and RIKEN approved the study, and all parents gave written informed consent.
Markers and genotyping
Microsatellite markers from the ABI Linkage Mapping Set MD-10 (Applied Biosystems, Foster City, CA, USA) were used for screening, and genetic mapping information on these markers was obtained from the web site of the Whitehead Institute (http://www.broad.mit.edu/cgi-bin/contig/phys_map). The allele frequencies and heterozygosity of these markers in the Japanese population have already been described (Ikari et al. 2001). Markers whose heterozygosity has been determined as less than 0.60 in the Japanese population were substituted with alternative ones. To avoid the problem of population stratification, data analyses were conducted with allele frequencies of the markers in founders of the sib pairs. The total number of markers was 399 and the average interval 9.9 cM. Polymerase chain reactions (PCRs) for genotyping were performed by the method described previously (Ikari et al. 2001). Pooled PCR products were mixed with GeneScan 500 LIZ Size Standard, and electrophoresis was performed on ABI 3700 capillary DNA sequencers (Applied Biosystems) according to the manufacturer’s protocol. Genotyping for each individual was performed using ABI GeneScan 3.5.2 and Genotyper 3.7 software (Applied Biosystems). Inconsistency within families was ruled out using the Checkfam program (http://www.genstat.net/checkfam/index.cgi?lang=ja).
Detailed mapping of a locus in 12q24
To obtain more information on a locus around the linkage peak in 12q24, we introduced three markers, one already known and two newly identified (Table 1). Primers used for amplification of these markers were as follows: D12S366F 5′-AAATACAGAGAATCTGGCTCG-3′, D12S366R 5′-GGCCGATCACTTCTTGAATC-3′, 12qMS054F 5′-CAATCCAGGAACCCAAGTAG-3′, 12qMS054R 5′-ACAAGAGACTGCAACCAGAG-3′, 12qMS069F 5′-ATCCAGGTATCCAGGAAAGG-3′, 12qMS069R 5′-GAAGGCAGATTTCCCTCAAC-3′.
Statistical analysis
The power of affected sib pair test was calculated using the previously described method (Risch 1990). Both single and multipoint nonparametric linkage (NPL) analyses for all genotype data was performed using the MAPMAKER/SIBS 2.0 program (Kruglyak and Lander 1995). Calculations of the identity by descent (IBD) distribution were conducted every 1.0 cM in the multipoint analyses. In the analyses, family members who had no obvious history of KD were treated as unknown as to whether they were affected. The program was also used to estimate information content. NPL scores and P values were calculated with the GENEHUNTER 2.1 program (Kruglyak et al. 1996); the sex-linked mode of MAPMAKER/SIBS and GENEHUNTER 1.3 program were used for the analysis of chromosome X.
Results
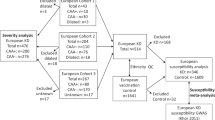
A total of 79 families including 75 full sib pairs, three sib trios, and one half sib pair were collected; samples from the half sib pair were not employed in this study. In 48 families, samples of either or both parents were available. Samples from unaffected siblings were also obtained in ten families (Table 2). The geographical distribution of patients recruited in this study as affected sib pairs is displayed in Fig. 1. The result of the power calculation of sib pair analysis is summarized in Table 3. According to this result, detection of suggestive linkage could be expected at loci with λs larger than 3.0 with an acceptable power (> 0.80). Average genotyping success rate was 98.4%, and the overall average of information content was 0.70.
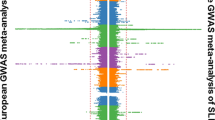
The results of the genomewide linkage analysis are displayed in Fig. 2. Ten chromosomal regions, 4q35, 5q34, 6q27, 7p15, 8q24, 12q24, 18q23, 19q13, Xp22, and Xq27 showed maximum lod scores (MLS) greater than 1.0 (Fig. 2a–d; Table 4). Among them, the most significant region was 12q24 (MLS 2.69, NPL 2.64, P=0.0043). The 1-lod support interval for this region spanned 25 cM (129 ∼ 154 cM). One hundred and twenty-eight genes are currently mapped within this interval (Table 5). Among them, 90 genes are believed to be expressed in organs related to immune function. Replacement and addition of markers around the peak of chromosome 12 increased information content from 0.66 to 0.84. In this condition, MLS of the locus was 2.26 and was still above the threshold of suggestive linkage (Fig. 3).
Genomewide linkage analysis of Kawasaki disease (KD). Genotype data for 78 Japanese KD families were analyzed by MAPMAKER/SIBS. The Y axis indicates the maximum lod score (MLS) values, and the X axis indicates the relative position of the microsatellite markers in each chromosome. The right side of the X axis corresponds to the q terminal of the chromosome
Information content and lod score with alternative marker set in 12q region. Markers of the first row are the set used in the initial scan. Replaced or added markers are in the second row. Plots for information content (IC) and maximum lod score (MLS) are drawn on the upper and lower part of the chart, respectively. Broken and continuous lines indicate the results in the initial scan and alternative high information scan, respectively
Discussion
KD is basically a self-limiting febrile illness that mainly affects infants and children. The most plausible trigger of KD, as determined from clinical features, epidemiologic profiles, and laboratory findings, would be some unknown infection(s). Several candidate infectious agents have been isolated from specimens of KD patients in the acute phase (Tsurumizu et al. 1991; Leung et al. 1993), but to date, no agents have been confirmed by the subsequent studies.
Since 1967, when KD was first reported by Kawasaki in Japan (Kawasaki 1967), much effort has been directed toward clarification of the pathogenesis and pathophysiology of the disease, but no therapeutic or preventive strategy based on etiological evidence has been developed. On the other hand, epidemiologic studies have shed light on the genetic aspects of KD as a result of two findings. One is an ethnic difference in disease prevalence: in Hawaii, increased prevalence in the Japanese American population has been observed (Dean et al. 1982). The other is familial aggregation of the disease: the relative risk of siblings (λs) for KD is about ten, and two-generation KD patients have been observed more frequently than expected (Fujita et al. 1989; Uehara et al. 2003). These findings clearly indicate the existence of some genetic factor(s) responsible for susceptibility to the disease. However, no multigenerational family suggesting a distinct inheritance pattern has been reported during the almost 40 years since KD was identified. Hence, we employed an NPL method in this study. As shown in Table 3, the observed linkage peaks were almost concordant using the two analysis methods. However, we considered that the utmost care was needed in the interpretation of NPL scores in this study. In contrast to the affected sib pair analysis, the affection status of all relatives analyzed is reflected in the NPL score. Although correct information from the parents of their history of KD in childhood is essential, no such record was available. Thus, it is possible that unrecognized patients exist in the parents and the NPL score was underestimated.
The strongest evidence of linkage was observed in the 12q24 region (MLS=2.69, NPL=2.64, P=0.0043). The level of likelihood of the region surpassed the threshold of suggestive linkage for genomewide sib pair analysis of complex trait (lod=2.2) (Lander and Kruglyak 1995). To achieve precise calculation of shared alleles identical by descent (IBD) in sib pair analysis, both collection of as many parents’ samples as possible and usage of highly informative markers are important. In this study, we obtained samples of at least one parent from 48 pedigrees. In addition, the information content of the linkage peak on 12q24 was improved by addition and replacement of the markers.
Interestingly, positive linkage results have been reported in several studies on allergic disorders on the very same chromosomal region (Shao et al. 2004; Yokouchi et al. 2002; CSGA 1997). Together with the fact that children with KD history are at higher risk for allergy (Matsubara et al. 1998), it is plausible that the responsible gene(s) exists in this loci. As demonstrated in Table 5, more than a hundred known and predicted genes are located in the 1-lod support interval in this region. We are now searching for the gene variation for KD susceptibility by use of gene expression and linkage disequilibrium data available from public databases.
In terms of KD susceptibility, a significant association between a single nucleotide polymorphisms (SNP) in the IL-4 gene and KD was reported by Burns et al. (2005). IL-4 is known to be located in the 5q31.1 region, where the so-called cytokine gene cluster exists. Several diseases related to autoimmunity or allergy have already been mapped to this region (Rioux et al. 2001; Palmer et al. 2001). In this study, although not significant, nominal evidence of linkage was observed in the region (MLS=0.98, data not shown). IL-12B, located on 5q33.3 region, is one of the most-studied genes as candidates for immune-mediated diseases. In our study, another linkage peak was observed in the 5q33-34 region (Table 3). It is possible that variation of the genes associated with such disorders is associated with marked activation of the immune system in acute-phase KD.
Another chromosomal region that has frequently been investigated in relation to immune disorders is 6p21, where human leukocyte antigen (HLA) genes are located. The association between HLA and development of KD remains controversial (Barron et al. 1992; Fildes et al. 1992; Kaslow et al. 1985; Krensky et al. 1981, 1983; Matsuda et al. 1977). No evidence of linkage was observed in the 6p21 region in the present study, which might indicate that HLA does not play a major role in terms of KD susceptibility.
Development of a prior prediction method of disease severity and outcome for individual KD patients in the acute phase would be a significant contribution to clinical practice. Responsiveness to immunoglobulin therapy and/or vascular wall fragility to immune insults could be also determined by genetic factors that could predict disease outcome, if clarified. Applying a positional candidate gene approach to the preliminary results of this linkage study, we have already identified an SNP within the CD40 ligand gene on chromosome X that is associated with coronary artery lesions in male KD patients (Onouchi et al. 2004). Although only a few concordant sib pair cases of coronary artery lesions were involved in the present study, other linkage peaks observed may also represent loci for susceptibility to cardiac complication.
The present study has revealed ten regions potentially involved in KD susceptibility. This is the first genome-wide genetic analysis for KD. Our findings will facilitate identification of genetic variations associated with KD susceptibility. The accumulation of knowledge on genetic factors should clarify etiology and pathogenesis of the disease and lead to the development of effective therapeutic measures.
References
Barron KS, Silverman ED, Gonzales JC, St Clair M, Anderson K, Reveille JD (1992) Major histocompatibility complex class II alleles in Kawasaki syndrome—lack of consistent correlation with disease or cardiac involvement. J Rheumatol 19:1790–1793
Burns JC (2002) Commentary: translation of Dr. Tomisaku Kawasaki’s original report of fifty patients in 1967. Pediatr Infect Dis J 21:993–995
Burns JC, Shimizu C, Shike H, Newburger JW, Sundel RP, Baker AL, Matsubara T, Ishikawa Y, Brophy VA, Cheng S, Grow MA, Steiner LL, Kono N, Cantor RM. (2005) Family-based association analysis implicates IL-4 in susceptibility to Kawasaki disease. Genes Immun 6:438–444
Dean AG., Melish ME, Hicks R, Palumbo NE (1982) An epidemic of Kawasaki syndrome in Hawaii. J Pediatr 100:552–557
Fildes N, Burns JC, Newburger JW, Klitz W, Begovich AB (1992) The HLA class II region and susceptibility to Kawasaki disease. Tissue Antigens 39:99–101
Fujita Y, Nakamura Y, Sakata K, Hara N, Kobayashi M, Nagai M, Yanagawa H, Kawasaki T (1989) Kawasaki disease in families. Pediatrics 84:666–669
Ikari K, Onda H, Furushima K, Maeda S, Harata S, Takeda J (2001) Establishment of an optimized set of 406 microsatellite markers covering the whole genome for the Japanese population. J Hum Genet 46:207–210
Kaslow RA, Bailowitz A, Lin FY, Koslowe P, Simonis T, Israel E (1985) Association of epidemic Kawasaki syndrome with the HLA-A2, B44, Cw5 antigen combination. Arthritis Rheum 28:938–940
Kato H, Koike S, Yamamoto M, Ito Y, Yano E (1975) Coronary aneurysms in infants and young children with acute febrile mucocutaneous lymph node syndrome. J Pediatr 86:892–898
Kato H, Sugimura T, Akagi T, Sato N, Hashino K, Maeno Y, Kazue T, Eto G, Yamakawa R (1996) Long-term consequences of Kawasaki disease. A 10- to 21-year follow-up study of 594 patients. Circulation 94:1379–1385
Kawasaki T (1967) Acute febrile mucocutaneous syndrome with lymphoid involvement with specific desquamation of the fingers and toes: my clinical observation (in Japanese). Jpn J Allergy 16:178–222
Krensky AM, Berenberg W, Shanley K, Yunis EJ (1981) HLA antigens in mucocutaneous lymph node syndrome in New England. Pediatrics 67:741–743
Krensky AM, Grady S, Shanley KM, Berenberg W, Yunis EJ (1983) Epidemic and endemic HLA-B and DR associations in mucocutaneous lymph node syndrome. Hum Immunol 6:75–77
Kruglyak L, Lander ES (1995) Complete multipoint sib-pair analysis of qualitative and quantitative traits. Am J Hum Genet 57:439–454
Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363
Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247
Leung DY, Meissner HC, Fulton DR, Murray DL, Kotzin BL, Schlievert PM (1993) Toxic shock syndrome toxin-secreting Staphylococcus aureus in Kawasaki syndrome. Lancet 342:1385–1388
Matsubara T, Fujita Y, Sato T, Sasai K, Furukawa S (1998) The prevalence of allergy in Kawasaki disease. Allergy 53:815–816
Matsuda I, Hattori S, Nagata N, Fruse A, Nambu H (1977) HLA antigens in mucocutaneous lymph node syndrome. Am J Dis Child 131:1417–1418
Onouchi Y, Onoue S, Tamari M, Wakui K, Fukushima Y, Yashiro M, Nakamura Y, Yanagawa H, Kishi F, Ouchi K, Terai M, Hamamoto K, Kudo F, Aotsuka H, Sato Y, Nariai A, Kaburagi Y, Miura M, Saji T, Kawasaki T, Nakamura Y, Hata A (2004) CD40 ligand gene and Kawasaki disease. Eur J Hum Genet 12:1062–1068
Palmer LJ, Barnes KC, Burton PR, Chen H, Cookson WO, Collaborative Study on the Genetics of Asthma, Deichmann KA, Elston RC, Holloway JW, Jacobs KB, Laitinen T, Wjst M (2001) Meta-analysis for linkage to asthma and atopy in the chromosome 5q31–33 candidate region. Hum Mol Genet 10:891–899
Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, Kocher K, Miller K, Guschwan S. Kulbokas EJ, O’Leary S, Winchester E, Dewar K, Green T, Stone V, Chow C, Cohen A, Langelier D, Lapointe G, Gaudet D, Faith J, Branco N, Bull SB, McLeod RS, Griffiths AM, Bitton A, Greenberg GR, Lander ES, Siminovitch KA, Hudson TJ (2001) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet 29:223–228
Risch N (1990) Linkage strategies for genetically complex traits. II. The power of affected relative pairs. Am J Hum Genet 46:229–241
Shao C, Suzuki Y, Kamada F, Kanno K, Tamari M, Hasegawa K, Aoki Y, Kure S, Yang X, Endo H, Takayanagi R, Nakazawa C, Morikawa T, Morikawa M, Miyabayashi S, Chiba Y, Karahashi M, Saito S, Tamura G, Shirakawa T, Matsubara Y (2004) Linkage and association of childhood asthma with the chromosome 12 genes. J Hum Genet 49:115–122
The Collaborative Study on the Genetics of Asthma (CSGA) (1997) A genome-wide search for asthma susceptibility loci in ethnically diverse populations. Nat Genet 15:389–392
Tsurumizu T, Okonogi H, Shibusawa T, Hashimoto T, Makino M, Ota H, Kurosaki T, Toba T (1991) A case of Kawasaki’s disease combined with septicemia–isolation of Streptococcus sanguis (MCLS-1) and Streptococcus pyogenes from blood at the acute stage. Kansenshogaku Zasshi (Jpn) 65:124–128
Uehara R, Yashiro M, Nakamura Y, Yanagawa H (2003) Kawasaki disease in parents and children. Acta Paediatr 92:694–697
Yanagawa H, Nakamura Y, Yashiro M, Ojima T, Tanihara S, Oki I, Zhang T (1998) Results of the nationwide epidemiologic survey of Kawasaki disease in 1995 and 1996 in Japan. Pediatrics 102:E65
Yokouchi Y, Shibasaki M, Noguchi E, Nakayama J, Ohtsuki T, Kamioka M, Yamakawa-Kobayashi K, Ito S, Takeda K, Ichikawa K, Nukaga Y, Matsui A, Hamaguchi H, Arinami T (2002) A genome-wide linkage analysis of orchard grass-sensitive childhood seasonal allergic rhinitis in Japanese families. Genes Immun 3:9–13
Acknowledgments
We thank patients and their families for participating in the project. We are grateful to Dr. Etsuko Tsuda, Dr. Shigeo Kojima, Dr. Marina Takenaka, Dr. Yutaka Izumi, Dr. Zenshiro Onouchi, Dr. Kaname Koyama, Dr. Atushi Ono, Dr. Fumie Furukawa, Dr. Takuji Otsuka, Dr. Takashi Hashimoto, Dr. Reiko Yabuta, Dr. Hirotoshi Iwahori, Dr. Yujiro Kimura, Dr. Yuji Murata, Dr. Ken Suzaki, Dr. Ken Takagawa, Dr. Michindo Ninomiya, Dr. Yukinobu Shibata, Dr. Minoru Kubo, Dr. Yuriko Sekine, Dr. Eikan Koh, Dr. Takeshi Terao, Dr. Mayumi Sugimori, Dr. Takashi Soga, Dr. Shigeki Terada, Dr. Nobuyuki Kiyosawa, Dr. Kunihiro Hamamoto, Dr. Noriko Baba, Dr. Hideo, Yamaguchi, Dr. Tatsuo Handa, Dr. Masashi Shiomi, Dr., Kensuke Harada, Dr. Yoshiyasu Yamada, Dr. Kazue IInuma, Dr. Sumio Hyodo, Dr. Masaru Miura, Dr. Haruo Terada, Dr. Mayumi Onda, Dr. Yoshiharu Suzuki, Dr. Shiro Tsuchiya, Dr. Naoto Tanaka, Dr. Masaki Nakagawa, Dr. Kisei Minami, Dr. Atsushi Toyoda, Dr. Chieko Udo, Dr. Kenji Yabuta, Dr. Tomio Kobayashi, Dr. Akira Kohno, Dr. Masumi Koujiro, Dr. Yasuhiko Tonegawa, Dr. Yuji Sakakura, Dr. Yutaka Yamada, Dr. Masanobu Ito, Dr. Fumiko Baba, Dr. Junji Suga, Dr. Kiyotaka Usui, Dr. Takashi Kawamura, Dr. Katsumi Takei, Dr. Seiji Noma, Dr. Mamie Watanabe, Kazunobu Ouchi, Dr. Tsutomu Saji, Dr. Tsuyoshi Isobe, and all their colleagues for cooperating in sample collection. The authors declare that they have no competing financial interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Onouchi, Y., Tamari, M., Takahashi, A. et al. A genomewide linkage analysis of Kawasaki disease: evidence for linkage to chromosome 12. J Hum Genet 52, 179–190 (2007). https://doi.org/10.1007/s10038-006-0092-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-006-0092-3
Keywords
This article is cited by
-
Immunogenetics of Kawasaki disease
Clinical Reviews in Allergy & Immunology (2020)
-
Simultaneous development of Kawasaki disease following acute human adenovirus infection in monozygotic twins: A case report
Pediatric Rheumatology (2017)
-
The Roles of Ca2+/NFAT Signaling Genes in Kawasaki Disease: Single- and Multiple-Risk Genetic Variants
Scientific Reports (2014)
-
ITPKC and CASP3 polymorphisms and risks for IVIG unresponsiveness and coronary artery lesion formation in Kawasaki disease
The Pharmacogenomics Journal (2013)
-
A genome-wide sib-pair linkage analysis of ossification of the posterior longitudinal ligament of the spine
Journal of Bone and Mineral Metabolism (2013)