Abstract
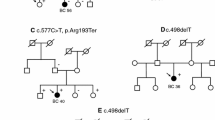
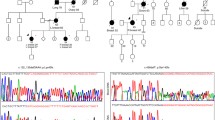
The RAD51C gene has been recently proposed as a high-penetrance breast and ovarian cancer gene. However, early replication studies have failed to confirm the finding. Thus, further studies in larger cohorts should be conducted in order to clarify the role of RAD51C as a cancer susceptibility gene. Here, we describe a high-resolution melting analysis (HRMA)-based method developed for presequence screening of RAD51C sequence variants. We have screened RAD51C sequence variants by HRMA in 492 breast cancer patients with family history of breast and/or ovarian cancer that were previously tested negative for BRCA1/2. All variants were confirmed by direct sequencing. We have detected 12 different RAD51C germ-line sequence variants, including eight transitions, two transversion, and two indels (insA, and delT). All these variants generated melting profiles which differ from wild type homozygous controls. Interestingly, we have identified one clearly pathogenic mutation (c.774delT) in the subset of 101 breast and ovarian cancer families, supporting that RAD51C is a human breast and ovarian cancer susceptibility gene.

Similar content being viewed by others
References
Gonzalez KD, Noltner KA, Buzin CH, Gu D, Wen-Fong CY, Nguyen VQ, Han JH, Lowstuter K, Longmate J, Sommer SS, Weitzel JN (2009) Beyond Li Fraumeni syndrome: clinical characteristics of families with p53 germline mutations. J Clin Oncol 27:1250–1256
Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R (1997) Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet 16:64–67
Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, Houben M, Crepin E, van Veghel-Plandsoen M, Elstrodt F, van Duijn C, Bartels C, Meijers C, Schutte M, McGuffog L, Thompson D, Easton D, Sodha N, Seal S, Barfoot R, Mangion J, Chang-Claude J, Eccles D, Eeles R, Evans DG, Houlston R, Murday V, Narod S, Peretz T, Peto J, Phelan C, Zhang HX, Szabo C, Devilee P, Goldgar D, Futreal PA, Nathanson KL, Weber B, Rahman N, Stratton MR (2002) CHEK2-Breast cancer consortium. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 31:55–59
Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, North B, McGuffog L, Evans DG, Eccles D, Breast Cancer Susceptibility Collaboration (UK), Easton DF, Stratton MR, Rahman N (2006) Truncating mutations in the Fanconi anemia J gene BRP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet 38:1239–1241
Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D, Breast Cancer Susceptibility Collaboration (UK), Easton DF, Stratton MR (2007) PALB2 which encodes a BRCA2-interacting protein is a breast cancer susceptibility gene. Nat Genet 39:165–167
Bergman A, Abel F, Behboudi A, Yhr M, Mattsson J, Svensson JH, Karlsson P, Nordling M (2008) No germline mutations in supposed tumour suppressor genes SAFB1, SAFB2 in familial breast cancer with linkage to 19p. BMC Med Genet 9:108
Oldenburg RA, Kroeze-Jansema KH, Houwing-Duistermaat JJ, Bayley JP, Dambrot C, van Asperen CJ, van den Ouweland AM, Bakker B, van Beers EH, Nederlof PM, Vasen H, Hoogerbrugge N, Cornelisse CJ, Meijers-Heijboer H, Devilee P (2008) Genome-wide linkage scan in Dutch hereditary non-BRCA1/2 breast cancer families identifies 9q21–22 as a putative breast cancer susceptibility locus. Genes Chromosomes Cancer 47:947–956
Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, Fraternali F, Freund M, Hartmann L, Grimwade D, Roberts RG, Schaal H, Mohammed S, Rahman N, Schindler D, Mathew CG (2010) Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet 42:406–409
Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, Freund M, Lichtner P, Hartmann L, Schaal H, Ramser J, Honisch E, Kubisch C, Wichmann HE, Kast K, Deissler H, Engel C, Müller-Myhsok B, Neveling K, Kiechle M, Mathew CG, Schindler D, Schmutzler RK, Hanenberg H (2010) Germline mutations in breast, ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 42:410–414
Zheng Y, Zhang J, Hope K, Niu Q, Huo D, Olopade OI (2010) Screening RAD51C nucleotide alterations in patients with a family history of breast, ovarian cancer. Breast Cancer Res Treat 124:857–861
Akbari MR, Tonin P, Foulkes WD, Ghadirian P, Tischkowitz M, Narod SA (2010) RAD51C germline mutations in breast, ovarian cancer patients. Breast Cancer Res 12:404
Weitzel JN, Lagos VI, Cullinane CA, Gambol PJ, Culver JO, Blazer KR, Palomares MR, Lowstuter KJ, MacDonald DJ (2007) Limited family structure, BRCA gene mutation status in single cases of breast cancer. JAMA 297:2587–2595
Díez O, Osorio A, Durán M, Martinez-Ferrandis JI, De la Hoya M, Salazar R, Vega A, Campos B, Rodríguez-López R, Velasco E, Chaves J, Díaz-Rubio E, JesúsCruz J, Torres M, Esteban E, Cervantes A, Alonso C, San Román JM, González-Sarmiento R, Miner C, Carracedo A, Eugenia Armengod M, Caldés T, Benítez J, Baiget M (2003) Analysis of BRCA1 and BRCA2 genes in Spanish breast/ovarian cancer patients: a high proportion of mutations unique to Spain and evidence of founder effects. Hum Mutat 22:301
Gutiérrez-Enríquez S, de la Hoya M, Martínez-Bouzas C, Sanchez de Abajo A, Ramón y Cajal T, Llort G, Blanco I, Beristain E, Díaz-Rubio E, Alonso C, Tejada MI, Caldés T, Diez O (2007) Screening for large rearrangements of the BRCA2 gene in Spanish families with breast/ovarian cancer. Breast Cancer Res Treat 103:103–107
De la Hoya M, Gutiérrez-Enríquez S, Velasco E, Osorio A, de Sanchez Abajo A, Vega A, Salazar R, Esteban E, Llort G, Gonzalez-Sarmiento R, Carracedo A, Benítez J, Miner C, Díez O, Díaz-Rubio E, Caldes T (2006) Genomic rearrangements at the BRCA1 locus in Spanish families with breast/ovarian cáncer. Clin Chem 52:1480–1485
Rozen Steve, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Vossen RH, Aten E, Roos A, den Dunnen JT (2009) High-resolution melting analysis (HRMA): more than just sequence variant screening. Hum Mutat 30:860–866
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C (2009) Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acid Res 37:e67
Krypuy M, Ahmed AA, Etemadmoghadam D, Hyland SJ, Australian Ovarian Cancer Study Group, De Fazio A, Fox SB, Brenton JD, Bowtell DD, Dobrovic A (2007) High resolution melting for mutation scanning of TP53 exons 5–8. BMC Cancer 7:168
Bastien R, Lewis TB, Hawkes JE, Quackenbush JF, Robbins TC, Palazzo J, Perou CM, Bernard PS (2008) High-throughput amplicon scanning of the TP53 gene in breast cancer using high-resolution fluorescent melting curve analyses and automatic mutation calling. Hum Mutat 29:757–764
Millat G, Chanavat V, Rodriguez-Lafrasse C, Rousson R (2009) Rapid sensitive and inexpensive detection of SCN5A genetic variations by high resolution melting analysis. Clin Biochem 42:491–499
Takano EA, Mitchell G, Fox SB, Dobrovic A (2008) Rapid detection of carriers with BRCA1 and BRCA2 mutations using high resolution melting analysis. BMC Cancer 8:59
Acknowledgments
We wish to thank the patients and their families for their cooperation. This study was supported by research grants FIS 09/00859 and RTICC 06/0020/0021, Instituto de Salud Carlos III (FEDER), Spanish Ministry of Science and Innovation. The study was also supported by Fundacion Mutua Madrileña research grant FMMA-08.
Conflict of interests
None.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Romero, A., Pérez-Segura, P., Tosar, A. et al. A HRM-based screening method detects RAD51C germ-line deleterious mutations in Spanish breast and ovarian cancer families. Breast Cancer Res Treat 129, 939–946 (2011). https://doi.org/10.1007/s10549-011-1543-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-011-1543-x