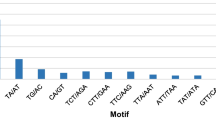
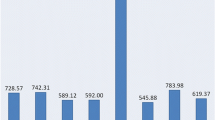
Abstract
The European hazelnut (Corylus avellana L.) is one of the most important nut crops. In this work, we characterize functional microsatellite or simple sequence repeat (SSR) markers for genetic analysis and molecular breeding in this species. A total of 38,454 Betulaceae EST sequences from NCBI resulted in 1,282 non-redundant EST-SSRs. Dinucleotide repeats were the most abundant (63.9 %), followed by trinucleotides (33.8 %). The putative functions of the non-redundant EST-SSRs were classified according to gene ontology (GO) categories (biological process, molecular function, and cellular component). A total of 921 sequences showed significant hits with the non-redundant protein database, and GO categories were assigned to 696 (75.5 %) of them. Flanking primer pairs were designed for 78 di- and trinucleotide EST-SSRs from Alnus glutinosa L. (29), Betula pendula Roth (26), and Betula platyphylla Suckaczev (23). Further, 41 dinucleotide repeats selected from hazelnut transcriptome sequences were added. Thirty-six out 119 primer pairs generated amplification products in six hazelnut accessions and in the samples of the species from which they were isolated. Among them, 20 were polymorphic when tested on 18 hazelnut cultivars. Fifteen loci are suitable for mapping in a F1 population of ‘Tonda Gentile delle Langhe’ × ‘Merveille de Bollwiller’ and 11 of them were functionally annotated. The cross-species transferability of 36 EST-SSR loci within nine Corylus species was also performed. The success rate of markers transferability (excluding C. avellana) ranged from 11 to 100 %, with an average of 55 %. The EST-SSRs developed increase the number of markers currently available for hazelnut.

Similar content being viewed by others
References
Acuña CV, Fernandez P, Villalba PV, García MN, Hopp HE, Marcucci Poltri SN (2012) Discovery, validation, and in silico functional characterization of EST-SSR markers in Eucalyptus globulus. Tree Genet Genomes 8:289–301
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H et al (2000) Gene ontology: tool for the unification of biology. Gene Ontol Consort Nat Genet 25:25–29
Bacchetta L, Aramini M, Zini A, Di Giammatteo V, Spera D, Drogoudi P, Rovira M, Silva AP, Solar A, Botta R (2013) Fatty acids and alpha-tocopherol composition in hazelnut (Corylus avellana L.): a chemometric approach to emphasize the quality of European germplasm. Euphytica 191:57–73
Bassil NV, Botta R, Mehlenbacher SA (2005a) Microsatellite markers in the hazelnut: isolation, characterization and cross-species amplification in Corylus. J Am Soc Hort Sci 130:543–549
Bassil NV, Botta R, Mehlenbacher SA (2005b) Additional microsatellites of the European hazelnut. Acta Hort 686:105–110
Bassil NV, Boccacci P, Botta R, Postman J, Mehlenbacher SA (2013) Nuclear and chloroplast microsatellite markers to assess genetic diversity and evolution in hazelnut species, hybrids and cultivars. Genet Resour Crop Evol 60:543–568
Beltramo C, Boccacci P, Sandoval Prando MA, Portis E, Botta R (2014) Development of a genetic linkage map in hazelnut (Corylus avellana L.) for the detection of QTLs. Acta Hort 1052:99–104
Boccacci P, Botta R (2010) Microsatellite variability and genetic structure in hazelnut (Corylus avellana L.) cultivars from different growing regions. Sci Hortic 124:128–133
Boccacci P, Akkak A, Bassil NV, Mehlenbacher SA, Botta R (2005) Characterization and evaluation of microsatellite loci in European hazelnut (Corylus avellana L.) and their transferability to other Corylus species. Mol Ecol Notes 5:934–937
Boccacci P, Akkak A, Botta R (2006) DNA-typing and genetic relationships among European hazelnut (Corylus avellana L.) cultivars using microsatellite markers. Genome 49:598–611
Boccacci P, Rovira M, Botta R (2008) Genetic diversity of hazelnut (Corylus avellana L.) germplasm in northeastern Spain. HortScience 43:667–672
Boccacci P, Aramini M, Valentini N, Bacchetta L, Rovira M, Drogoudi P, Silva AP, Solar A, Calizzano F, Erdoğan V, Cristofori V, Ciarmiello LF, Contessa C, Ferreira JJ, Marra FP, Botta R (2013) Molecular and morphological diversity of on-farm hazelnut (Corylus avellana L.) landraces from southern Europe and their role in the origin and diffusion of cultivated germplasm. Tree Genet Genomes 9:1465–1480
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Campa A, Trabanco E, Pérez-Vega E, Rovira M, Ferreira JJ (2011) Genetic relationship between cultivated and wild hazelnuts (Corylus avellana L.) collected in northern Spain. Plant Breed 130:360–366
Chen ZD, Manchester SR, Sun HY (1999) Phylogeny and evolution of the Betulaceae as inferred from DNA sequences, morphology, and paleobotany. Am J Bot 86:1168–1181
Chen C, Zhou P, Choi YA, Huang S, Gmitter FG Jr (2006) Mining and characterizing microsatellites from Citrus ESTs. Theor Appl Genet 112:1248–1257
Conesa A, Götz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Decroocq V, Fave MG, Hagen L, Bordenave L, Decroocq S (2003) Development and transferability of apricot and grape EST microsatellite markers across taxa. Theor Appl Genet 106:912–922
Edwards KJ, Barker JHA, Daly A, Jones C, Karp A (1996) Microsatellite libraries enriched for several microsatellite sequences in plants. BioTechiques 20:758–760
Erdoğan V, Mehlenbacher SA (2000) Phylogenetic relationships of Corylus species (Betulaceae) based on nuclear ribosomal DNA ITS region and chloroplast matK gene sequences. Syst Bot 25:727
FAOstat (2014) Agriculture data. http://faostat.fao.org/site/339/default.aspx. Accessed 26 Jan 2014
Feng SP, Li WG, Huang HS, Wang JY, Wu YT (2009) Development, characterization and cross-species/genera transferability of EST-SSR markers for rubber tree (Hevea brasiliensis). Mol Breed 23:85–97
Fraser LG, Harvey CF, Crowhurst RN, De Silva HN (2004) EST derived microsatellites from Actinidia species and their potential for mapping. Theor Appl Genet 108:1010–1016
Gasic K, Han Y, Kertbundit S, Shulaev V, Iezzoni AF, Stover EW, Bell RL, Wisniewski ME, Korban SS (2009) Characteristics and transferability of new apple EST-derived SSRs to other Rosaceae species. Mol Breed 23:397–411
Gökirmak T, Mehlenbacher SA, Bassil NV (2009) Characterization of European hazelnut (Corylus avellana L.) cultivars using SSR markers. Genet Resour Crop Evol 56:147–172
Gürcan K, Mehlenbacher SA (2010a) Transferability of microsatellite markers in the Betulaceae. J Am Soc Hort Sci 135:159–173
Gürcan K, Mehlenbacher SA (2010b) Development of microsatellite marker loci for European hazelnut (Corylus avellana L.) from ISSR fragments. Mol Breed 26:551–559
Gürcan K, Mehlenbacher SA, Botta R, Boccacci P (2010a) Development, characterization, segregation, and mapping of microsatellite markers for European hazelnut (Corylus avellana L.) from enriched genomic libraries and usefulness in genetic diversity studies. Tree Genet Genomes 6:513–531
Gürcan K, Mehlenbacher SA, Erdoğan V (2010b) Genetic diversity in hazelnut (Corylus avellana L.) cultivars from Black Sea countries assessed using SSR markers. Plant Breed 129:422–434
Heath MC (2000) Hypersensitive response-related death. Plant Mol Biol 44:321–334
Heesacker A, Kishore VK, Gao W, Tang S, Kolkman JM, Gingle A, Matvienko M, Kozik A, Michelmore RM, Lai Z, Rieseberg LH, Knapp SJ (2008) SSRs and INDELs mined from the sunflower EST database: abundance, polymorphisms, and cross-taxa utility. Theor Appl Genet 117:1021–1029
Huang H, Lu J, Hunter W, Dowd S, Dang P (2011) Mining and validating grape (Vitis L.) ESTs to develop EST-SSR markers for genotyping and mapping. Mol Breed 28:241–254
Järvinen P, Palmé A, Morales LO, Lännenpää M, Keinänen M, Sopanen T, Lascoux M (2004) Phylogenetic relationships of Betula species (Betulaceae) based on nuclear ADH and chloroplast matK sequences. Am J Bot 91:1834–1845
Kurepa J, Smalle JA (2008) Structure, function and regulation of plant proteasomes. Biochimie 90:324–335
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Mehlenbacher SA, Brown RN, Nouhra EN, Gökirmak T, Bassil NV, Kubisiak TL (2006) A genetic linkage map for hazelnut (Corylus avellana L.) based on RAPD and SSR markers. Genome 49:122–133
Metzgar D, Bytof J, Wills C (2000) Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res 10:72–80
Mukhopadhyay A, Vij S, Tyagi AK (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc Natl Acad Sci USA 101:6309–6314
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Rowley ER, Fox SE, Bryant DW, Sullivan CM, Priest HD, Givan SA, Mehlenbacher SA, Mockler TC (2012) Assembly and characterization of the European hazelnut ‘Jefferson’ transcriptome. Crop Sci 52:2679–2686
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Sathuvalli VR, Mehlenbacher SA (2012) Characterization of American hazelnut (Corylus americana) accessions and Corylus americana × Corylus avellana hybrids using microsatellite markers. Genet Resour Crop Evol 59:1055–1075
Sathuvalli VR, Chen H, Mehlenbacher SA, Smith DC (2011) DNA markers linked to eastern filbert blight resistance in ‘Ratoli’ hazelnut (Corylus avellana L.). Tree Genet Genomes 7:337–345
Thiel T, Michalek W, Varshney R, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Thomas MR, Matsumoto S, Cain P, Scott NS (1993) Repetitive DNA of grapevine: classes present and sequences suitable for cultivar identification. Theor Appl Genet 86:173–180
Tsutsui T, Asada Y, Tamaoki M, Ikeda A, Yamaguchi J (2008) Arabidopsis CAD1 negatively controls plant immunity mediated by both salicylic acid-dependent and-independent signalling pathways. Plant Sci 175:604–611
Varshney R, Granner A, Sorrells M (2005) Genic microsatellite markers in plants: features and applications. Trends Biotech 23:48–55
Victoria FC, da Maia LC, de Oliveira AC (2011) In silico comparative analysis of SSR markers in plants. BMC Plant Biol 11:15
Whitcher IN, Wen J (2001) Phylogeny and biogeography of Corylus (Betulaceae): inference from ITS sequences. Syst Bot 26:283–298
Wöhrmann T, Weising K (2011) In silico mining for simple sequence repeat loci in a pineapple expressed sequence tag database and cross-species amplification of EST-SSR markers across Bromeliaceae. Theor Appl Genet 123:635–647
Yoo K-O, Wen J (2002) Phylogeny and biogeography of Carpinus and subfamily Coryloideae (Betulaceae). Int J Plant Sci 163:641–650
Zhang LS, Becquet V, Li SH, Zhang D (2003) Optimization of multiplex PCR and multiplex gel electrophoresis in sunflower SSR analysis using infrared fluorescence and tailed primers. Acta Bot Sin 45:1312–1318
Acknowledgments
The research was funded by the Foundation Cassa di Risparmio di Torino (CRT).
Author information
Authors and Affiliations
Corresponding author
Additional information
P. Boccacci and C. Beltramo have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Boccacci, P., Beltramo, C., Sandoval Prando, M.A. et al. In silico mining, characterization and cross-species transferability of EST-SSR markers for European hazelnut (Corylus avellana L.). Mol Breeding 35, 21 (2015). https://doi.org/10.1007/s11032-015-0195-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0195-7