Abstract
Due to spectral overlap, the number of fluorescent labels for imaging cryomicrotome detection was limited to 4. The aim of this study was to increase the separation of fluorescent labels. In the new imaging cryomicrotome, the sample is cut in slices of 40 μm. Six images are taken for each cutting plane. Correction for spectral overlap is based on linear combinations of fluorescent images. Locations of microspheres are determined by using the system point spread function. Five differently colored microspheres were injected in vivo distributed over two major coronaries, the left anterior descending and left circumflex artery. Under absence of collateral flow, microspheres outside of target perfusion territories were not found and the procedure did not generate false positive detection when spectral overlap was relevant. In silico-generated microspheres were used to test the effect of background image, transparency correction, and color separation. The percentage of microspheres undetected was 2.3 ± 0.8% in the presence and 1.5 ± 0.4% in the absence of background structures with a density of 900 microspheres per color per cm3. The image analysis method presented here, allows for an increased number of experimental conditions that can be investigated in studies of regional myocardial perfusion.
Similar content being viewed by others
1 Introduction
Labeled microspheres have been used for more than half a century to quantify perfusion distribution, either over the circulatory system as a whole or within an organ [14]. In this method, labeled microspheres are injected into the circulation either as a bolus or by continuous infusion over a period of time [12]. The working principle is that these microspheres, typically with a diameter around 15 μm, travel with the blood stream and become trapped in the microcirculation since they cannot pass the capillaries. Ultimately, they shift from the capillary lumen to the interstitium [4, 10]. The number of microspheres in a certain region, relative to the total amount of injected microspheres and weighted by the blood flow at the site of injection, represents the regional perfusion in volume flow per unit mass.
Several methods have been developed permitting the distinction of different labels. The classical labels were radioactive [3], colored [9] or following recent development, fluorescent [8]. Tissue samples for measurements were originally obtained by cutting the tissue in the area of interest into pieces of 0.1–2 cm3 depending on the study aim [1]. The radioactivity in these samples was then measured directly [6] or these were processed to determine the total fluorescence or number of microspheres [17].
In a recently introduced and more advanced method, an imaging cryomicrotome is used that allows the position of individual microspheres to be detected [11]. In this method, the tissue block under investigation is frozen and sliced with a cryomicrotome at a preset thickness, e.g., 40 μm at a time. After each cut, fluorescent images are taken with a high-resolution digital camera. Image processing then yields the x–y–z coordinates of the microspheres [2, 11].
The classical and cryomicrotome method share a commonality in that the tissue examined is destroyed. The information retained in the classical method has a spatial resolution of the tissue sample size which has an under limit of 0.1 g, while in the cryomicrotome the resolution is of the voxel size of the camera and slice thickness, which is a factor 1 × 106 higher. Moreover, the region of interest for calculation of microsphere density can be selected at will in the cryomicrotome method but not in the classical methods. In addition, injection of fluorescent cast material into the vasculature prior to freezing allows detection of the arterial tree structures down to small arterioles with a diameter of 20–40 μm [15, 16]. Hence, the imaging cryomicrotome enables the combined measurements of blood vessel structure and flow distribution within an organ. However, until now, only four different fluorescent labels could be distinguished with the imaging cryomicrotome method [11], which is a limitation compared to the earlier radioactive labeled methods where up to nine different labels could be detected.
The purpose of this study was to increase the number of differently labeled fluorescents. We present a method to improve the fluorescent color separation by correcting for spectral overlap based on a linear combination of the images. Our approach permits differentiation of six fluorescent labels, of which five are used for the microspheres and one for the infused casting material. The proposed method was validated both in silico and with actual data obtained from canine hearts.
2 Materials and methods
2.1 Canine dataset
Datasets for this study were obtained from three animals of a series of physiological experiments carried out in canines according to a protocol that was approved by the Institutional Animal Care and Use Committee of the University of Utrecht Medical Center. After pre-medication with Dormitor 0.03 mg/kg i.m. (intra-muscular), Ketamine 0.03 mg/kg i.m. and Atropine 0.5 mg i.m. anesthesia was induced by Sufenta forte 10 μg/kg/h i.v. the animals were intubated, and a surgical level of anesthesia was maintained by mechanical ventilation with a mixture of Propofol 24 mg/kg/h i.v., Sufenta forte 3 μg/kg/h i.v. and oxygen. The hearts were exposed by left thoractomy via the fourth intercostal space. Under fluoroscopic imaging, 8F guiding catheters were positioned at the ostium of the left circumflex artery, LCX, and the left anterior descending artery, LAD. Microspheres were gradually infused over a period of 1 min (standard infusion pump, Harvard Apparatus, USA). The microsphere dose for each infusion was approximately 1.0 × 105 (15μm, Invitrogen, The Netherlands) as determined from microsphere concentration in the storage vial and volume withdrawn. The colors used (with corresponding excitation and emission maxima in nm between brackets) were: green (454/492), yellow (518/521), red (570/598), carmine (582/614) and scarlet (651/680). Microspheres were injected at different physiological conditions such as baseline and hyperemia. However, for this study, it was not attempted to correlate distributions to these physiological conditions.
After the experiment, the animal was killed by fibrillating the heart after which it was excised, and the right and left main coronary arteries were cannulated and perfused with a buffered solution containing adenosine for vasodilation of the microcirculation. Subsequently, the main vessels were infused with cast replica material (Batson no. 17, Polysciences, USA) consisting of a monomer base solution, a catalyst and a promoter as described previously [15, 16]. Potomac Yellow (440/490) (Radiant Colour, Belgium) provided the fluorescent base for the replica plastic. The cast material was allowed to harden over a period of 24 h at ambient temperature. The fully prepared heart was embedded in carboxymethylcellulose sodium solvent 5% (Brunschwig Chemie, The Netherlands) and Indian ink 5% (Royal Talens, The Netherlands), frozen at −20°C and placed in the cryomicrotome for subsequent cutting [15].
2.2 The imaging cryomicrotome
The frozen specimen was cut with the imaging cryomicrotome (Fig. 1) at 40 μm slice thickness. After each slice, the surface of the remaining bulk material was imaged using a 4000 × 4000 pixel digital camera (Apogee Alta U-16, USA) equipped with a variable focus lens (Nikon 70-180 mm, The Netherlands). The sample was illuminated with a 300 W Xenon lamp (Asmuth ASPE300BF, Germany). Filter wheels containing five optical filters each (Chroma Technology Corp., USA) were used to select specific bandwidths of light for excitation and emission. The filters were defined by the central wavelength and bandwidth in nm; for excitation: (A) 414:30, (B) 440:20, (C) 510:20, (D) 560:20 and (E) 640:20; and for the emission: (1) 480:20, (2) 505:30, (3) 555:30, (4) 635:30 and (5) 712:75. The combination of filter settings will further be referred to in terms of this alpha-numerical notation (Table 1).
2.3 Image processing
Three-dimensional (3D) computer reconstructions of the specimen were obtained based on 40 × 40 × 40 μm3 voxels. The image stack resulting from reflection imaging delivered an outline representation of the heart, while the stacks of fluorescent images yielded a 3D reconstruction of the vasculature and distributions of microspheres with different fluorescent labels. A series of algorithms was developed and applied to correct for imaging artifacts as described below.
2.4 Correction for transparency effects
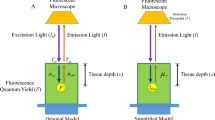
Without compensation, light coming from deeper layers of the sample introduces false detections of microspheres. The effect can span a depth of up to 200 μm and hence, the majority of microspheres visible at the surface are not actually present in the surface layer, slice n. Up until now, the cutting depth at which the light disappeared was taken as the location of the microsphere [2]. However, subsequent microspheres will be counted as one, which results into loss of observations. Therefore, we propose an image processing step to correct for transparency effects prior to detection.
Since the point spread function of the imaging system is known as well as the total intensity at slice n + 1, we can mimic the intensity contribution of microspheres contained in the deeper layers. We used the theoretical model specific to the cryomicrotome setup as proposed by Rolf et al. [13], which consists of three terms. The first represents Lambert–Beers law for incoming light with intensity I 0,
Here, μt is the attenuation coefficient and z the distance from the light source. The second term is the intensity component of a point source radiating in all directions;
Convolution of this expression with the lens term in the form of a Gaussian function yields
An approximation of Eq. 3 can be made based on observations of simulated data, by fitting a two-dimensional Gaussian function to simulation results as proposed by Rolf et al. [13]. Since the width of the Gaussian is depth dependent, the point source radiation term can be omitted if σ written in the form σ1 z + σ2. Furthermore, the excitation and emission term for the attenuation are combined into a single exponential term. This yields a simplified expression for the point spread function (PSF):
Application of Eq. 4 allows a longitudinal, in the z direction, deconvolution based on sequential slice correction to compensate for the transparency effects of the tissue. Let I n be the total intensity at slice n, for an arbitrary set of excitation and emission filters, I n can then be written in discrete form as:
with I ms n , the intensity contribution from microspheres currently at slice n and I tr n the intensity contributed by deeper laying microspheres that are visible due to the transparency of the tissue. To describe I tr n , we use the relationship
For intensity changes over a single slice, z=1, the PSF can even be further simplified to:
With σt a contraction of σ1 and σ2. Substitution into Eq. 6 yields:
The expression for the intensity contribution of the in-plane microspheres is given by:
Equation 9 is used to correct for the transparency effect throughout the stack of images.
2.5 Correction for spectral overlap
Since the microspheres have overlapping excitation and emission spectra, no unique filter combination is able to image just a single color when more than four colors are present. Figure 2 shows the overlapping emission spectra of the microspheres and the cast material (Potomac Yellow) used in this study, along with the emission filters 1–5 and their respective bandwidths. The excitation filters are chosen to maximize the intensity response per color while keeping the overlap to a minimum. By employing combinations of images obtained with different filters, the number of colors can be extended to five plus that of the casting material. For each combination of excitation and emission filters, two different colors are imaged. In general, if \( I_{n}^{\sigma } \left( {i, \, j, \, \lambda_{\text{ex}} , \, \lambda_{\text{em}} } \right) \) is an image taken with a specific excitation and emission filter combination, the intensity of the microsphere distribution \( I_{n}^{a} (i,j) \) in the image will be seen along with distribution \( I_{n}^{b} \left( {i, \, j} \right) \) due to the spectral overlap,
Here, α(λex, λem) and β(λex, λem) are scale factors that depend on the excitation and emission wavelengths, ranging between 0 and 1. The filters λex and λem are chosen such that α(λex, λem) ≈ 1. By taking an additional image \( I_{n}^{\tau } (i, \, j,\lambda_{\text{ex}}^{{^{'} }} ,\lambda_{\text{em}}^{{^{'} }} ) \) with filter settings \( \lambda_{\text{ex}}^{{^{'} }} \) and \( \lambda_{\text{em}}^{{^{'} }} \) chosen such that \( I_{n}^{b} \left( {i, \, j} \right) \) is visible and \( I_{n}^{a} \left( {i, \, j} \right) \) is absent, we obtain:
where \( I_{n}^{c} \left( {i, \, j} \right) \) is the intensity of the distribution caused by spectral overlap with respect to \( I_{n}^{b} \left( {i, \, j} \right) \). We arrive at the unique color \( I_{n}^{a} \left( {i, \, j} \right) \) by subtraction
The factor Ф is introduced to compensate for the difference in exposure time between \( I_{n}^{\sigma } \left( {i, \, j, \, \lambda_{\text{ex}} , \, \lambda_{\text{em}} } \right) \) and \( I_{n}^{\tau } (i, \, j,\lambda_{\text{ex}}^{{^{'} }} ,\lambda_{\text{em}}^{{^{'} }} ) \). The validity of this approach derives from the fact that the position of the microspheres is fixed for each image and that any negative value as a result of subtraction is set to zero intensity in the final image.The actual linear combinations to obtain unique colors per image are shown in Table 2, where α, β, γ, δ, and ε represent the scale factors.
2.6 Correction for auto-fluorescence
Cardiac tissue exhibits strong auto-fluorescence, especially at the excitation wavelength of 440 nm. To correct for this, a two-dimensional white Top-hat transformation of the data is performed. The Top-hat transform is a morphologic operation to separate small scale structures in the image from large intensity gradients [7]. Applying this filtering, the background intensity is suppressed, but the absolute peak value at the microsphere location is lowered as well, thereby increasing the signal-to-noise ratio. To compensate for the reduced intensity, an intensity scaling filter is introduced as
where α is a lower threshold, β an upper threshold, and \( I_{n}^{{^{'} }} (i,j) \) is the rescaled intensity. The lower threshold α is chosen to be above the level of background noise and the upper threshold β is defined by the maximum intensity observation in the microsphere data.
2.7 Microsphere detection
A binary representation of the data is created by applying a threshold to the images, based on the median value of the intensity for the stack. Each cluster of bright pixels that are connected is initially counted as one microsphere observation. The observations are then checked for four criteria before the final microsphere location is accepted and stored.
According to the first criterion, the average intensity of the pixels in an observation must be above a user-defined threshold to ensure that any reminiscent light due to transparency artifacts is not identified as a microsphere. Second, we apply the criterion that observations must be convex in plane. This follows from the intensity profile of the in-plane microspheres that can be estimated by a Gaussian profile [13]. By stepping along the contour of the observation, the sequence of steps needed to return to the starting point can be evaluated for the x and y directions. If a sign change occurs along the sequence more than twice for a direction, the observation is characterized as non-convex and is eliminated, as illustrated in Fig. 3. The third criterion identifies saddle points in the observation that occur since microspheres may form clusters. The clustered microspheres may be counted as one when not corrected for. If the test for a saddle point evaluates to true, the individual peaks in intensity within the connected region above the median value of the stack are stored as microspheres locations. Finally, the fourth criterion identifies large intensity observations with a single peak, which can result from two microspheres that are too close together to be distinguishable due to the limits of the optical setup. These are separated and are stored as two adjacent observations.
Only if the four criteria are passed, the 2D location of the identified microsphere in the current image is stored in a text file for further processing.
2.8 In silico validation
In order to evaluate the robustness of the spectral overlap correction, the full detection method for all five colors of microspheres along with the cast material was additionally tested with a simulated dataset. The parameters to create the test datasets were chosen such that they corresponded to actual in vivo measurements as measured by Rolf et al. We used Eq. 4 to produce the datasets, with varying attenuation coefficients, in-plane and depth-dependent standard deviation for the Gaussian kernel and microsphere density. For σ1, we chose a fixed value of 0.1 pixel, σ2 ranged from 0.8 for B2, 1.0 for C3, 1.2 for B4/D4 and 1.4 pixel for E5 and the attenuation coefficient ranged in pixels from 0.5 for B2, 0.4 for C3, 0.3 for B4/D4 and 0.2 for E5 accordingly. The microspheres were randomly distributed in the in silico datasets, although in vivo this is typically not the case [5]. To overcome this limitation, we created two distributions with different densities; a high density set, with approximately 900 microspheres/cm3 to resemble local high flow regions and a density stack of approximately 500 microspheres/cm3 comparable to the average number of microspheres/cm3 as observed in animal experiments. The individual microspheres within the in silico distributions were assigned different intensity levels between 100 and 255. This is in accordance with in vivo observations where the intensity can vary due to the position of the microsphere in the current slice, variation of intensity of microspheres and regional tissue properties. Finally, a realistic background image was added by superimposing in vivo images onto the in silico datasets as a real life representation of the variation in background intensity due to the camera properties and the surrounding tissue. These images were analyzed with the corrections as described above.
3 Results
3.1 Transparency effects and auto-fluorescence
A typical example of transparency effects is shown in Fig. 4. These images show a cropped region with injected microspheres in the left ventricular wall in two successive slides. In general, most of the microsphere observations in the top surface slice originate from deeper layers, which necessitates compensation for the transparency effect. Figure 5 demonstrates the success of the proposed method for eliminating the transparency effects. Three microspheres are shown in a 3D subset that belong to the same canine data set as Fig. 4. The transparency effects are completely corrected for this subset and the microspheres are only visible in the x–y plane which is the basis for the subsequent detection algorithm.
Illustration of incorrect microsphere detection due to transparency effects. A cropped region of 250 × 250 pixels is shown for two sequential slices (a before b) containing only carmine microspheres. The small arrow indicates a microsphere that was only visible in slice (a) and would be counted correctly. The large arrow shows a cluster of microspheres not actually present in either slice (a) or (b), but still visible due to the transparency of the surrounding tissue material, possibly leading to false identifications
The effect of transparency for three single microspheres shown in a 3D representation of a cropped volume of 15 × 30 × 30 pixels. a Without correction; b with correction for transparency effects using Eq. 9
The effect of Top-hat filtering and intensity scaling on suppressing the auto-fluorescence is illustrated in Fig. 6 for a single microsphere in the canine dataset. The image was taken with an excitation filter 440 nm and emission filter 635 nm. A circle with the radius in the same size as the imaged microspheres was applied for the structural element of the Top-hat filter. However, depending on the local background the intensity of the microsphere is reduced by this operation. The scaling filter is applied to linearly stretch the intensity for the new background to the old peak value. The result of the filter and scaling operations is presented for a line as well as a cropped 2D region of the real data.
Effect of Top-hat filter and intensity rescaling on a cropped region of 60 × 60 pixels. a Microsphere with simulated auto-fluorescent light in the form of an intensity gradient. b 1D intensity profile across the midline in the raw image, before and after top-hat filtering (dark grey). c Top-hat filtered intensity profile after rescaling with a lower threshold of 20. d 2D image result of these operations
3.2 Microsphere counts and location
3.2.1 In silico dataset
The simulated images allowed us to validate the detection of microspheres as well as the performance of the algorithm in the presence of microsphere clusters.
Quantitative agreement for colored microsphere detection in the in silico datasets is reported in Table 3 for a high (900 microspheres/cm3) and low concentration (500 microspheres/cm3), respectively. The count number for both density simulations was checked manually to prevent a false positive detection cancelling out a false negative detection. In order to validate the correction of the cast material in the green microsphere images, actual cast images were added to the simulated dataset for the case of green microspheres. For both the groups, the detection accuracy was greater than 98% without noise and only slightly affected by noise, resulting in a reduction of accuracy in the order of 0.9%.
3.2.2 In vivo dataset
In two of the three hearts, microspheres with overlapping emission spectra were injected into the LAD and LCX respectively. The combinations were red into the LAD and yellow into the LCX for one heart (Fig. 7a) and scarlet into the LAD and carmine into the LCX for the other heart (Fig. 7b). The color distributions are plotted in the transverse plane for a tissue slab of 10 mm thickness. In Fig. 7a, a downsampled representation of the cast material is also visualized along with the microsphere distributions. Please note that no false positives are generated in either color.
Separation of microsphere colors, shown as 3D microsphere distributions for a 10 mm thick slab in the sagittal (left panels) and transverse (right panels) plane of two canine hearts. a, b Yellow microspheres injected into the LCX and red microspheres into the LAD, together with detected fluorescent casting material. c, d Carmine microspheres injected into the LCX and scarlet microspheres into the LAD. A clear separation of perfusion territories can be seen for both hearts. Note: These figures do not display the actual microsphere colors, but are pseudo-colored to emphasize the distinction between the different perfusion territories
From each of these colors, saddle points and high intensity detections were evaluated. The percentage of detected saddle points (indicating doubles) and high intensities (clusters of microspheres) amounted to a mean value and standard deviation of 0.75 ± 0.53 and 1.81 ± 0.85%, respectively. In addition to the microsphere injections with spectral overlap, a third color of microspheres (green) was injected into the LAD that had no spectral overlap with the prior injections. Scarlet microspheres combined with yellow microspheres were also injected into the LAD of the third heart. This was done to evaluate the amount of clustering for differently labeled microspheres without spectral overlap. For all the three hearts, the number of such clusters was determined and resulted in a mean of 0.26% with a standard deviation of 0.15%.
4 Discussion
We presented a method to correct for spectral overlap of fluorescent emission light, which allows the use and successful separation of a larger number of fluorescent labels in imaging cryomicrotome applications. With our image processing algorithms, we successfully distinguished one color for the casting material and an additional five colors for microspheres, thereby increasing the number of fluorescent colors successfully detected from 4 to 6. An increase in labels is important, since it enlarges the maximum number of experimental conditions that can be investigated in a single animal which in turn lowers the number of animal experiments needed for a particular protocol and reduces the organ-to-organ variability across animals.
The in silico simulations demonstrated that over 98% of the microspheres were correctly counted for a realistic range of microsphere densities. The number of missed counts was only 1.5 ± 0.4% for high microsphere density (900 spheres/cm3) and decreased to 0.9 ± 0.8% for a lower concentration (500 spheres/cm3). With background images added, the number of missed microspheres increased to 2.3 ± 0.8 and 1.8 ± 1.3%, respectively. The error was not specific for any color.
The data obtained from frozen canine hearts showed a clear demarcation zone between the LAD and LCX perfusion territories indicating the absence of false positive detection of microspheres. Although, the number of saddle points and high intensity observations was very low, it increased with the wavelength of the emission light. This is due to the widening of the point spread function as a consequence of the lower attenuation coefficient of the tissue for longer wavelengths in combination with the chromatic aberration of the lens system.
4.1 Factors influencing microsphere detection
The unwanted contribution of light due to spectral overlap is already minimal for selective excitation and emission filter combinations, hence β(λex, λem) ≪ α(λex, λem) (Eq. 10). For the casting material (Potomac Yellow), the single filter B1 (440/480) is sufficient, since the emission extends further into the near ultraviolet range, with no overlap up to 490 nm. For the other colors, the value for the scale factor ε (Eq. 12) typically ranges from 0.5 to 1. Especially for the red and carmine microspheres, β(λex, λem) ≈ α(λex, λem) as the overlap in emission spectra is clearly discernible (Fig. 2). Hence, spectral overlap correction is a necessity for these colors.
In our image analysis, we assumed that the point spread function is not a function of spatial coordinates. However, in reality, this is not the case and hence transparency correction is complicated due to the anisotropic optical properties of the tissue. To avoid the problem related to anisotropy, we used an underestimate for the attenuation coefficient as proposed by Rolf et al. [13]. If the sequential slice correction is then applied, transparency may be overcorrected but this results not in undetected microspheres.
Dark current of the camera is a common source of error when imaging with exposure times in the order of seconds and adds to image degradation. It is the result of thermionic emission or other effects when there is no light incident on the charged coupled device and results in relatively bright pixels compared to the median value of all pixels in the image. The effects of this dark current are suppressed by active cooling of the camera, but may remain present to some degree as a consequence of long exposure times. Since the errors due to dark current are consistent in the longitudinal direction of the stack, the applied correction for transparency effects also removes potential dark current errors. The effect of dark current in isolated pixels on the detection of microspheres is further diminished, since a microsphere is classified by two or more pixels.
4.2 Clustering
Clustering of microspheres may additionally lead to incorrect microsphere identification. One may classify clustering into three types: (1) microspheres of the same color are clustered prior to injection, (2) single microspheres of the same color cluster together during the injection in the same blood vessel, and (3) microspheres of different colors cluster together during injection in the same blood vessel. Clustering of the same color as in possibilities 1 and 2 is detected by the saddle point checks and the identification of observations larger than two microspheres. Clustering of two microspheres of different colors, however, gives rise to possible loss of observations when there is spectral overlap, since part of the intensity belonging to a color of interest is removed. The clustering of originally detached microspheres by accidentally following the same path to the point of lodging as defined in possibilities 2 and 3 were addressed in silico and in vivo. In the simulated datasets, 3D distribution of microspheres were stochastically distributed. In only 1.5% of the voxels containing two microspheres, more than two were found. When counting the colors without spectral overlap injected into identical perfusion territories, on average only 0.26% of the total number of microspheres shared identical xyz coordinates.
4.3 Comparison with other methods of microsphere density estimation
The imaging cryomicrotome has several advantages over the more classical methods of microsphere density determination, where first the tissue has to be partitioned and then each tissue sample has to be processed to either count the microspheres by a flow cytometric quantification method or by determining the average tracer concentration. For the latter method, a correction is needed to account for the natural decay of fluorescence intensity or radioactivity, depending on the tracer used. This obviously introduces additional sources of error. This problem does not exist with the imaging cryomicrotome, since the microspheres are detected by the shape of corresponding fluorescence patterns rather than by intensity alone. The cytometric quantification method is also free of the intensity decay problem, but the resolution is not improved. With the datasets obtained from a cryomicrotome, any desired 3D analysis of microsphere distribution can be applied, since the spatial relation of the dataset with respect to the outline images is not destroyed. Moreover, in combination with the casting procedure, a detailed analysis of microsphere distribution (flow) with respect to the intramural vascular structure can be performed in organs such as the heart.
5 Conclusions
We demonstrated the feasibility of using different filter combinations to distinguish five separate microsphere colors in addition to the fluorescently labeled vascular tree. This allows high-resolution spatial measurements of local perfusion for five different experimental conditions in the same organ that can additionally be closely related to the detected vascular structure.
References
Austin RE Jr, Hauck WW, Aldea GS, Flynn AE, Coggins DL, Hoffman JI (1989) Quantitating error in blood flow measurements with radioactive microspheres. Am J Physiol 257:H280–H288
Bernard SL, Ewen JR, Barlow CH, Kelly JJ, McKinney S, Frazer DA, Glenny RW (2000) High spatial resolution measurements of organ blood flow in small laboratory animals. Am J Physiol Heart Circ Physiol 279:H2043–H2052
Buckberg GD, Luck JC, Payne DB, Hoffman JI, Archie JP, Fixler DE (1971) Some sources of error in measuring regional blood flow with radioactive microspheres. J Appl Physiol 31:598–604
Consigny PM, Verrier ED, Payne BD, Edelist G, Jester J, Baer RW, Vlahakes GJ, Hoffman JI (1982) Acute and chronic microsphere loss from canine left ventricular myocardium. Am J Physiol 242:H392–H404
Decking UK, Pai VM, Bennett E, Taylor JL, Fingas CD, Zanger K, Wen H, Balaban RS (2004) High-resolution imaging reveals a limit in spatial resolution of blood flow measurements by microspheres. Am J Physiol Heart Circ Physiol 287:H1132–H1140
Domenech RJ, Hoffman JI, Noble MI, Saunders KB, Henson JR, Subijanto S (1969) Total and regional coronary blood flow measured by radioactive microspheres in conscious and anesthetized dogs. Circ Res 25:581–596
Dougherty ER (1992) An introduction to morphological image processing. SPIE Optical Engineering Press, Bellingham
Glenny RW, Bernard S, Brinkley M (1993) Validation of fluorescent-labeled microspheres for measurement of regional organ perfusion. J Appl Physiol 74:2585–2597
Hale SL, Vivaldi MT, Kloner RA (1986) Fluorescent microspheres: a new tool for visualization of ischemic myocardium in rats. Am J Physiol 251:H863–H868
Hales JR, Cliff WJ (1977) Direct observations of the behaviour of microspheres in microvasculature. Bibl Anat 15:87–91
Kelly JJ, Ewen JR, Bernard SL, Glenny RW, Barlow CH (2000) Regional blood flow measurements from fluorescent microsphere images using an imaging cryomicrotome. Rev Sci Instrum 71:228–234
McDevitt DG, Nies AS (1976) Simultaneous measurement of cardiac output and its distribution with microspheres in the rat. Cardiovasc Res 10:494–498
Rolf MP, ter Wee R, van Leeuwen TG, Spaan JA, Streekstra GJ (2008) Diameter measurement from images of fluorescent cylinders embedded in tissue. Med Biol Eng Comput 46:589–596
Rudolph AM, Heymann MA (1967) The circulation of the fetus in utero methods for studying distribution of blood flow, cardiac output and organ blood flow. Circ Res 21:163–184
Spaan JA, ter Wee R, van Teeffelen JW, Streekstra G, Siebes M, Kolyva C, Vink H, Fokkema DS, VanBavel E (2005) Visualisation of intramural coronary vasculature by an imaging cryomicrotome suggests compartmentalisation of myocardial perfusion areas. Med Biol Eng Comput 43:431–435
van den Wijngaard JP, van Horssen P, ter Wee R, Coronel R, de Bakker JM, de Jonge N, Siebes M, Spaan JA (2010) Organization and collateralization of a subendocardial plexus in end-stage human heart failure. Am J Physiol Heart Circ Physiol 298:H158–H162
Van Oosterhout MF, Willigers HM, Reneman RS, Prinzen FW (1995) Fluorescent microspheres to measure organ perfusion: validation of a simplified sample processing technique. Am J Physiol 269:H725–H733
Acknowledgments
The authors would like to thank J. Weda (Dept. of Biomedical Engineering and Physics) for his assistance in obtaining the spectra of the fluorescent labels presented in this work. This work has been supported in part by The Netherlands Heart Foundation (NHS 2000.082, NHS2006B226, NHS 2006B186), by The Netherlands Organization for Scientific Research (ZONMW 91105008), and by the European Community’s Seventh Framework Program Grant FP7/2007-2013 under grant agreement no. 224495 (euHeart project).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
van Horssen, P., Siebes, M., Hoefer, I. et al. Improved detection of fluorescently labeled microspheres and vessel architecture with an imaging cryomicrotome. Med Biol Eng Comput 48, 735–744 (2010). https://doi.org/10.1007/s11517-010-0652-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11517-010-0652-8