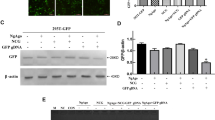
Abstract
The RNA-guided endonuclease clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (Cas9) derived from CRISPR systems is a simple and efficient genome-editing technology applied to various cell types and organisms. So far, the extensive approach to detect the cleavage activity of customized Cas9/guide RNA (gRNA) is T7 endonuclease I (T7EI) assay, which is time and labor consuming. In this study, we developed a visualized fluorescent reporter system to detect the specificity and cleavage activity of gRNA. Two gRNAs were designed to target porcine immunoglobulin M and nephrosis 1 genes. The cleavage activity was measured by using the traditional homology-directed repair (HDR)-based fluorescent reporter and the single-strand annealing (SSA)-based fluorescent reporter we established in this study. Compared with the HDR assay, the SSA-based fluorescent reporter approach was a more efficient and dependable strategy for testing the cleavage activity of Cas9/gRNA, thereby providing a universal and efficient approach for the application of CRISPR/Cas9 in generating gene-modified cells and organisms.






Similar content being viewed by others
References
Beumer, K. J., et al. (2008). Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proceedings of the National Academy of Sciences of the United States of America, 105(50), 19821–6.
Geurts, A. M., et al. (2009). Knockout rats via embryo microinjection of zinc-finger nucleases. Science, 325(5939), 433.
Joung, J. K., & Sander, J. D. (2013). TALENs: a widely applicable technology for targeted genome editing. Nature Reviews Molecular Cell Biology, 14(1), 49–55.
Miller, J. C., et al. (2011). A TALE nuclease architecture for efficient genome editing. Nature Biotechnology, 29(2), 143–8.
Li, D., et al. (2013). Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nature Biotechnology, 31(8), 681–3.
Li, W., et al. (2013). Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nature Biotechnology, 31(8), 684–6.
Jinek, M., et al. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337(6096), 816–21.
Mali, P., et al. (2013). RNA-guided human genome engineering via Cas9. Science, 339(6121), 823–6.
Ran, F. A., et al. (2013). Genome engineering using the CRISPR-Cas9 system. Nature Protocols, 8(11), 2281–308.
Wang, H., et al. (2013). One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell, 153(4), 910–8.
Chang, N., et al. (2013). Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Research, 23(4), 465–72.
Jao, L. E., Wente, S. R., & Chen, W. (2013). Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences of the United States of America, 110(34), 13904–9.
Hwang, W. Y., et al. (2013). Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PloS One, 8(7), e68708.
Yang, D., et al. (2014). Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. Journal of Molecular Cell Biology, 6(1), 97–9.
Yan, Q., et al. (2014). Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen (Lond), 3(1), 12.
Niu, Y., et al. (2014). Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell, 156(4), 836–43.
Friedland, A. E., et al. (2013). Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nature Methods, 10(8), 741–3.
Xie, K., & Yang, Y. (2013). RNA-guided genome editing in plants using a CRISPR-Cas system. Molecular Plant, 6(6), 1975–83.
Kim, H. J., et al. (2009). Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Research, 19(7), 1279–88.
Kim, H., et al. (2011). Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nature Methods, 8(11), 941–3.
Wilson, K. A., Chateau, M. L., & Porteus, M. H. (2013). Design and development of artificial zinc finger transcription factors and zinc finger nucleases to the hTERT Locus. Molecular Therapy Nucleic Acids, 2, e87.
Rouet, P., Smih, F., & Jasin, M. (1994). Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America, 91(13), 6064–8.
Garcia Civera, R., et al. (1980). Retrograde P wave polarity in reciprocating tachycardia utilizing lateral bypass tracts. European Heart Journal, 1(2), 137–45.
Segal, D. J., & Carroll, D. (1995). Endonuclease-induced, targeted homologous extrachromosomal recombination in Xenopus oocytes. Proceedings of the National Academy of Sciences of the United States of America, 92(3), 806–10.
Mali, P., Esvelt, K. M., & Church, G. M. (2013). Cas9 as a versatile tool for engineering biology. Nature Methods, 10(10), 957–63.
Fu, Y., et al. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nature Biotechnology, 32(3), 279–84.
Liu, Y., et al. (2014). A modified TALEN-based strategy for rapidly and efficiently generating knockout mice for kidney development studies. PloS One, 9(1), e84893.
Zhou, Y., et al. (2016) Enhanced genome editing in mammalian cells with a modified dual-fluorescent surrogate system. Cell Mol Life Sci.
Zou, J., et al. (2009). Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell, 5(1), 97–110.
Xiao, A., et al. (2014) CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31101781, 31072102) and the Programs Foundation of Ministry of Education of China (20110061110081).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Additional information
Yi Yang and Songcai Liu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yang, Y., Liu, S., Cheng, Y. et al. Highly Efficient and Rapid Detection of the Cleavage Activity of Cas9/gRNA via a Fluorescent Reporter. Appl Biochem Biotechnol 180, 655–667 (2016). https://doi.org/10.1007/s12010-016-2122-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2122-8