Abstract
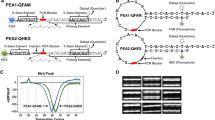
The tetra-primer amplification refractory mutation system–polymerase chain (ARMS–PCR) reaction is a simple and economical method to genotype single-nucleotide polymorphisms (SNPs). It uses four primers in a single PCR and is followed just by gel electrophoresis. However, the optimization step can be very hardworking and time-consuming. Hence, we propose to demonstrate and discuss critical steps for its development, in a way to provide useful information. Two SNPs that provided different amplification conditions were selected. DNA extraction methods, annealing temperatures, PCR cycles protocols, reagents, and primers concentration were also analyzed. The use of tetra-primer ARMS–PCR could be impaired for SNPs in DNA regions rich in cytosine and guanine and for samples with DNA not purified. The melting temperature was considered the factor of greater interference. However, small changes in the reagents concentration significantly affect the PCR, especially MgCl2. Balancing the inner primers band is also a key step. So, in order to balance the inner primers band, intensity is important to observe which one has the weakest band and promote its band by increasing its concentration. The use of tetra-primer ARMS–PCR attends the expectations of modern genomic research and allows the study of SNPs in a fast, reliable, and low-cost way.





Similar content being viewed by others
References
Collins, F. S., Morgan, M., & Patrinos, A. (2003). The Human Genome Project: lessons from large-scale biology. Science, 300(5617), 286–290.
The International HapMap Consortium. (2005). A haplotype map of the human genome. Nature, 437(7063), 1299–1320.
Shi, J., Wang, Y., & Huang, W. (2009). Development and application of genotyping technologies. Science in China, Series C: Life Sciences, 52(1), 17–23.
Shastry, B. S. (2002). SNP alleles in human disease and evolution. Journal of Human Genetics, 47(11), 561–566.
Rocha, D., Gut, I., Jeffereys, A. J., Kwok, P. Y., Brookes, A. J., & Chanock, S. J. (2006). Seventh international meeting on single nucleotide polymorphism and complex genome analysis: ever bigger scans and an increasingly variable genome. Human Genetics, 119(4), 451–456.
Manolio, T. A., Brooks, L. D., & Collins, F. S. A. (2008). HapMap harvest of insights into the genetics of common disease. Journal of Clinical Investigation, 118(5), 1590–1605.
Bui, M., & Liu, Z. (2009). Simple allele-discriminating PCR for cost-effective and rapid genotyping and mapping. Plant Methods, 5, 1. doi:10.1186/1746-4811-5-1.
Feero, W. G., Guttmacher, A. E., & Collins, F. S. (2010). Genomic medicine—an updated primer. New England Journal of Medicine, 362(21), 2001–2011.
Wang, W. P., Ni, K. Y., & Zhou, G. H. (2006). Approaches for SNP genotyping. Yi Chuan, 28(1), 117–126.
Shen, R., Fan, J. B., Campbell, D., Chang, W., Chen, J., Doucet, D., et al. (2005). High-throughput SNP genotyping on universal bead arrays. Mutation Research, 573(1–2), 70–82.
Griffin, T. J., & Smith, L. M. (2000). Single-nucleotide polymorphism analysis by MALDI-TOF mass spectrometry. Trends in Biotechnology, 18(2), 77–84.
Ahmadian, A., Ehn, M., & Hober, S. (2006). Pyrosequencing: History, biochemistry and future. Clinica Chimica Acta, 363(1–2), 83–94.
Syvänen, A. C. (2005). Toward genome wide SNP genotyping. Nature Genetics, 37(Suppl), S5–S10.
Chuang, L. Y., Yang, C. H., Tsui, K. H., Cheng, Y. H., Chang, P. L., Wen, C. H., et al. (2008). Restriction enzyme mining for SNPs in genomes. Anticancer Research, 28(4), 2001–2007.
Hamajima, N., Saito, T., Matsuo, K., & Tajima, K. (2002). Competitive amplification and unspecific amplification in polymerase chain reaction with confronting two-pair primers. Journal of Molecular Diagnostics, 4(2), 103–107.
Ye, S., Dhillon, S., Ke, X., Collins, A. R., & Day, I. N. (2001). An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Research, 29, e88. doi:10.1093/nar/29.17.e88.
Kwok, P. Y. (2001). Methods for genotype single nucleotide polymorphism. Annual Review of Genomics and Human Genetics, 2, 235–258.
Newton, C. R., Graham, A., Heptinstall, L. E., Powell, S. J., Summers, C., Kalsheker, N., et al. (1989). Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Research, 17(7), 2503–2516.
Little, S. (2001). Amplification-refractory mutation system (ARMS) analysis of point mutations. Current protocols Human Genet, vol 9.8 (Wiley Online Library): (pp. 1–12).
Wangkumhang, P., Chaichoompu, K., Ngamphiw, C., Ruangrit, U., Chanprasert, J., Assawamakin, A., et al. (2007). WASP: a web-based allele-Specific PCR assay design tool decting SNPs and mutations. BMC Genomics, 8, 275.
Rubio, M., Caranta, C., & Palloix, A. (2008). Functional markers for selection of potyvirus resistance alleles at the pvr2-eIF4E locus in pepper using tetra-primer ARMS-PCR. Genome, 51(9), 767–771.
Chiapparino, E., Lee, D., & Donini, P. (2004). Genotyping single nucleotide polymorphisms in barley tetra-primer ARMS–PCR. Genome, 47(2), 414–420.
Hubé, F., Reverdiau, P., Iochmann, S., & Gruel, Y. (2005). Improved PCR method for amplification of GC-rich DNA sequences. Molecular Biotechnology, 31(1), 81–84.
McDowell, D. C., Burns, N. A., & Parkes, H. C. (1998). Localised sequence regions possessing high melting temperatures prevent the amplification of a DNA mimic in competitive PCR. Nucleic Acids Resarch, 26(14), 3340–3347.
Henke, W., Herdel, K., Jung, K., Schnorr, D., & Loening, S. A. (1997). Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Research, 25(19), 3957–3958.
Varadaraj, K., & Skinner, D. M. (1994). Denaturants or cosolvents improve the specificity of PCR amplification of a G + C-rich DNA using genetically engineered DNA polymerases. Gene, 140(1), 1–5.
Garcés-Claver, A., Fellman, S. M., Gil-Ortega, R., Jahn, M., & Arnedo-Andrés, M. S. (2007). Identification, validation and survey of a single nucleotide polymorphism (SNP) associated with pungency in Capsicum spp. TAG Theoretical and Applied Genetics, 115(7), 907–916.
Markoulatos, P., Siafakas, N., & Moncany, M. (2002). Multiplex polymerase chain reaction: A practical approach. Journal of Clinical Laboratory Analysis, 16(1), 47–51.
Hohjoh, H., & Tokunaga, K. (2001). Allele-specific binding of the ubiquitous transcription factor OCT-1 to the functional single nucleotide polymorphism (SNP) sites in the tumor necrosis factor-alpha gene (TNFA) promoter. Genes and Immunity, 2(2), 105–109.
Baris, I., Etlik, O., Koksal, V., & Arican-Baris, S. T. (2010). Rapid diagnosis of spinal muscular atrophy using tetra-primer ARMS PCR assay: Simultaneous detection of SMN1 and SMN2 deletion. Molecular and Cellular Probes, 24(3), 138–141.
Chai, J., Xiong, Q., Zhang, P. P., Shang, Y. Y., Zheng, R., Peng, J., et al. (2010). Evidence for a new allele at the SERCA1 locus affecting pork meat quality in part through the imbalance of Ca2+ homeostasis. Molecular Biology Reports, 37(1), 613–619.
Acknowledgments
This work was supported by Fundação Hermínio Ometto/FHO and PIBIC/CNPq.
Conflict of interests
The authors declare that there are no conflicts of interest.
Ethical standard
This study was approved by the Committee of Ethic in Research and Scientific Merit of the Centro Universitário Herminio Ometto/UNIARARAS, number 744/2010.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Medrano, R.F.V., de Oliveira, C.A. Guidelines for the Tetra-Primer ARMS–PCR Technique Development. Mol Biotechnol 56, 599–608 (2014). https://doi.org/10.1007/s12033-014-9734-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-014-9734-4