Abstract
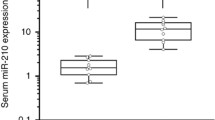
Glioma, the most common type of primary central nervous system cancers, was progressive with poor survival. MicroRNA, as a novel biomarker, was suspected to be novel biomarkers for glioma diagnosis and prognosis. The study aimed at investigating the diagnostic and predictive value of miR-221/222 family for glioma. In the first phase, we compared plasma miR-221/222 family levels between 50 glioma patients and 51 healthy controls by real-time qRT-PCR amplification. Meanwhile, a meta-analysis based on published studies and presents study was performed to explore the diagnostic performance of miR-221/222 family in human cancers. In the second phase, we correlated the miR-221/222 family expression level with prognosis of glioma using Kaplan-Meier survival curves. The plasma miR-221/222 family levels were found to be significantly upregulated in glioma patients (P = 0.001). The ROC curve analysis yielded an AUC values of 0.84 (95 % confidence interval (CI): 0.74–0.93) for miR-221 and 0.92 (95 % CI 0.87–0.94) for miR-222. In the meta-analysis, the summary receiver operating characteristic (sROC) was plotted with an AUC of 0.82 (95 % CI 0.78–0.85) for miR-221/222 family. It was also demonstrated that high positive plasma miR-221 and miR-222 were both correlated with poor survival rate (miR-221: HR = 2.13; 95 % CI, 1.05–4.31; miR-222: HR = 2.09; 95 % CI, 1.00–4.37). This study demonstrated that the detection of the miRNA-221/222 family should be considered as a new additional tool to better characterize glioma.





Similar content being viewed by others
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM (2010) Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer J int du Cancer 127(12):2893–2917. doi:10.1002/ijc.25516
Turner JD, Williamson R, Almefty KK, Nakaji P, Porter R, Tse V, Kalani MY (2010) The many roles of microRNAs in brain tumor biology. Neurosurg Focus 28(1):E3. doi:10.3171/2009.10.FOCUS09207
Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P (2007) The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 114(2):97–109. doi:10.1007/s00401-007-0243-4
Dolecek TA, Propp JM, Stroup NE, Kruchko C (2012) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro-Oncology 14(Suppl 5):v1–v49. doi:10.1093/neuonc/nos218
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359(5):492–507. doi:10.1056/NEJMra0708126
Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10(5):459–466. doi:10.1016/S1470-2045(09)70025-7
Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352(10):987–996. doi:10.1056/NEJMoa043330
Kuzniecky R, de la Sayette V, Ethier R, Melanson D, Andermann F, Berkovic S, Robitaille Y, Olivier A, Peters T, Feindel W (1987) Magnetic resonance imaging in temporal lobe epilepsy: pathological correlations. Ann Neurol 22(3):341–347. doi:10.1002/ana.410220310
Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, O’Fallon JR, Schaefer PL, Scheithauer BW, James CD, Buckner JC, Jenkins RB (2001) PTEN mutation, EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme. J Natl Cancer Inst 93(16):1246–1256
Wang SI, Puc J, Li J, Bruce JN, Cairns P, Sidransky D, Parsons R (1997) Somatic mutations of PTEN in glioblastoma multiforme. Cancer Res 57(19):4183–4186
Watanabe K, Tachibana O, Sata K, Yonekawa Y, Kleihues P, Ohgaki H (1996) Overexpression of the EGF receptor and p53 mutations are mutually exclusive in the evolution of primary and secondary glioblastomas. Brain Pathol (Zurich, Switzerland) 6(3):217–223, discussion 223–214
Duffield JS, Grafals M, Portilla D (2013) MicroRNAs are potential therapeutic targets in fibrosing kidney disease: lessons from animal models. Drug Discov Today Dis Model 10(3):e127–e135. doi:10.1016/j.ddmod.2012.08.004
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Cohen S (2010) MicroRNAs in animal development. Editorial. Semin Cell Dev Biol 21(7):727. doi:10.1016/j.semcdb.2010.07.004
Nana-Sinkam P, Croce CM (2010) MicroRNAs in diagnosis and prognosis in cancer: what does the future hold? Pharmacogenomics 11(5):667–669. doi:10.2217/pgs.10.57
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043):834–838. doi:10.1038/nature03702
Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K (2010) The microRNA spectrum in 12 body fluids. Clin Chem 56(11):1733–1741. doi:10.1373/clinchem.2010.147405
Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafre SA, Farace MG (2007) miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chemist 282(32):23716–23724. doi:10.1074/jbc.M701805200
Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G (2012) miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med 12(1):27–33
Waters PS, McDermott AM, Wall D, Heneghan HM, Miller N, Newell J, Kerin MJ, Dwyer RM (2012) Relationship between circulating and tissue microRNAs in a murine model of breast cancer. PLoS One 7(11):e50459. doi:10.1371/journal.pone.0050459
Mazeh H, Mizrahi I, Halle D, Ilyayev N, Stojadinovic A, Trink B, Mitrani-Rosenbaum S, Roistacher M, Ariel I, Eid A, Freund HR, Nissan A (2011) Development of a microRNA-based molecular assay for the detection of papillary thyroid carcinoma in aspiration biopsy samples. Thyroid Off J Am Thyroid Assoc 21(2):111–118. doi:10.1089/thy.2010.0356
Hu Z, Dong J, Wang LE, Ma H, Liu J, Zhao Y, Tang J, Chen X, Dai J, Wei Q, Zhang C, Shen H (2012) Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis 33(4):828–834. doi:10.1093/carcin/bgs030
Gombos K, Horvath R, Szele E, Juhasz K, Gocze K, Somlai K, Pajkos G, Ember I, Olasz L (2013) miRNA expression profiles of oral squamous cell carcinomas. Anticancer Res 33(4):1511–1517
Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, Gasparini P, Gonelli A, Costinean S, Acunzo M, Condorelli G, Croce CM (2009) miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16(6):498–509. doi:10.1016/j.ccr.2009.10.014
Zhang C, Kang C, You Y, Pu P, Yang W, Zhao P, Wang G, Zhang A, Jia Z, Han L, Jiang H (2009) Co-suppression of miR-221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo. Int J Oncol 34(6):1653–1660
Zhang J, Han L, Ge Y, Zhou X, Zhang A, Zhang C, Zhong Y, You Y, Pu P, Kang C (2010) miR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol 36(4):913–920
Bush VJ, Janu MR, Bathur F, Wells A, Dasgupta A (2001) Comparison of BD Vacutainer SST Plus Tubes with BD SST II Plus Tubes for common analytes. Clinica Chimica Acta Int J Clin Chemist 306(1–2):139–143
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327(7414):557–560. doi:10.1136/bmj.327.7414.557
Jackson D, White IR, Thompson SG (2010) Extending DerSimonian and Laird’s methodology to perform multivariate random effects meta-analyses. Stat Med 29(12):1282–1297. doi:10.1002/sim.3602
Deeks JJ, Macaskill P, Irwig L (2005) The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 58(9):882–893. doi:10.1016/j.jclinepi.2005.01.016
Guo HQ, Huang GL, Guo CC, Pu XX, Lin TY (2010) Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Dis Markers 29(5):251–258. doi:10.3233/dma-2010-0755
Pu XX, Huang GL, Guo HQ, Guo CC, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY (2010) Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol 25(10):1674–1680. doi:10.1111/j.1440-1746.2010.06417.x
Ryu JK, Matthaei H, Dal Molin M, Hong SM, Canto MI, Schulick RD, Wolfgang C, Goggins MG, Hruban RH, Cope L, Maitra A (2011) Elevated microRNA miR-21 levels in pancreatic cyst fluid are predictive of mucinous precursor lesions of ductal adenocarcinoma. Pancreatol Off J Int Assoc Pancreatol (IAP) [et al] 11(3):343–350. doi:10.1159/000329183
Wu CW, Dong YJ, Ng S, Leung WW, Wong CY, Sung JJ, Chan FKL, Yu J (2011) Identification of a panel of microRNAs in stool as screening markers for colorectal cancer. Gastroenterology 140(5):S73
Yaman Agaoglu F, Kovancilar M, Dizdar Y, Darendeliler E, Holdenrieder S, Dalay N, Gezer U (2011) Investigation of miR-21, miR-141, and miR-221 in blood circulation of patients with prostate cancer. Tumour Biol J Int Soc Oncodevelopmental Biol Med 32(3):583–588. doi:10.1007/s13277-011-0154-9
Pai R, Nehru GA, Samuel P, Selvan B, Kumar R, Jacob PM, Nair A (2012) Discriminating thyroid cancers from benign lesions based on differential expression of a limited set of miRNA using paraffin embedded tissues. Indian J Pathol Microbiol 55(2):158–162. doi:10.4103/0377-4929.97845
Song MY, Pan KF, Su HJ, Zhang L, Ma JL, Li JY, Yuasa Y, Kang D, Kim YS, You WC (2012) Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One 7(3):e33608. doi:10.1371/journal.pone.0033608
Wu Q, Wang C, Lu Z, Guo L, Ge Q (2012) Analysis of serum genome-wide microRNAs for breast cancer detection. Clinica Chimica Acta Int J Clin Chemist 413(13–14):1058–1065. doi:10.1016/j.cca.2012.02.016
Xie HT, Chu ZX, Wang H (2012) Serum microRNA expression profile as a biomarker in diagnosis of acute myeloid leukemia. J Clin Pediatr 30(05):421–424
Zaravinos A, Radojicic J, Lambrou GI, Volanis D, Delakas D, Stathopoulos EN, Spandidos DA (2012) Expression of miRNAs involved in angiogenesis, tumor cell proliferation, tumor suppressor inhibition, epithelial-mesenchymal transition and activation of metastasis in bladder cancer. J Urol 188(2):615–623. doi:10.1016/j.juro.2012.03.122
Cai H, Yuan Y, Hao YF, Guo TK, Wei X, Zhang YM (2013) Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med Oncol (Northwood, London, England) 30(1):452. doi:10.1007/s12032-012-0452-0
Srivastava A, Goldberger H, Dimtchev A, Ramalinga M, Chijioke J, Marian C, Oermann EK, Uhm S, Kim JS, Chen LN, Li X, Berry DL, Kallakury BV, Chauhan SC, Collins SP, Suy S, Kumar D (2013) MicroRNA profiling in prostate cancer—the diagnostic potential of urinary miR-205 and miR-214. PLoS One 8(10):e76994. doi:10.1371/journal.pone.0076994
Thapa DR, Hussain SK, Tran WC, D’Souza G, Bream JH, Achenback CJ, Ayyavoo V, Detels R, Martinez-Maza O (2014) Serum microRNAs in HIV-infected individuals as pre-diagnosis biomarkers for AIDS-NHL. J Acquir Immune Defic Syndr 66(2):229–237. doi:10.1097/qai.0000000000000146
Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM (2011) QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 155(8):529–536. doi:10.7326/0003-4819-155-8-201110180-00009
Sterne JA, Sutton AJ, Ioannidis JP, Terrin N, Jones DR, Lau J, Carpenter J, Rucker G, Harbord RM, Schmid CH, Tetzlaff J, Deeks JJ, Peters J, Macaskill P, Schwarzer G, Duval S, Altman DG, Moher D, Higgins JP (2011) Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 343:d4002. doi:10.1136/bmj.d4002
Burgoyne AM, Palomo JM, Phillips-Mason PJ, Burden-Gulley SM, Major DL, Zaremba A, Robinson S, Sloan AE, Vogelbaum MA, Miller RH, Brady-Kalnay SM (2009) PTPmu suppresses glioma cell migration and dispersal. Neuro-Oncology 11(6):767–778. doi:10.1215/15228517-2009-019
Chen L, Zhang J, Han L, Zhang A, Zhang C, Zheng Y, Jiang T, Pu P, Jiang C, Kang C (2012) Downregulation of miR-221/222 sensitizes glioma cells to temozolomide by regulating apoptosis independently of p53 status. Oncol Rep 27(3):854–860. doi:10.3892/or.2011.1535
Quintavalle C, Mangani D, Roscigno G, Romano G, Diaz-Lagares A, Iaboni M, Donnarumma E, Fiore D, De Marinis P, Soini Y, Esteller M, Condorelli G (2013) MiR-221/222 target the DNA methyltransferase MGMT in glioma cells. PLoS One 8(9):e74466. doi:10.1371/journal.pone.0074466
Wu C, Li M, Hu C, Duan H (2014) Prognostic role of microRNA polymorphisms in patients with advanced esophageal squamous cell carcinoma receiving platinum-based chemotherapy. Cancer Chemother Pharmacol 73(2):335–341. doi:10.1007/s00280-013-2364-x
Wang J, Liu S, Sun GP, Wang F, Zou YF, Jiao Y, Ning J, Xu J (2014) Prognostic significance of microRNA-221/222 expression in cancers: evidence from 1,204 subjects. Int J Biol Markers. doi:10.5301/jbm.5000058
Yang J, Zhang JY, Chen J, Xu Y, Song NH, Yin CJ (2014) Prognostic role of microRNA-221 in various human malignant neoplasms: a meta-analysis of 20 related studies. PLoS One 9(1):e87606. doi:10.1371/journal.pone.0087606
Acknowledgments
This work was supported by Science and Technology Development Project of Shandong Province, China (Grant No. 2014GGB14439 and 2014GGB14257).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, R., Pang, B., Xin, T. et al. Plasma miR-221/222 Family as Novel Descriptive and Prognostic Biomarkers for Glioma. Mol Neurobiol 53, 1452–1460 (2016). https://doi.org/10.1007/s12035-014-9079-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-014-9079-9