Abstract
Preterm delivery is associated with neurodevelopmental impairment caused by environmental and genetic factors. Dysfunction of the excitatory amino acid transporter 2 (EAAT2) and the resultant impaired glutamate uptake can lead to neurological disorders. In this study, we investigated the role of single nucleotide polymorphisms (SNPs; g.-200C>A and g.-181A>C) in the EAAT2 promoter in susceptibility to brain injury and neurodisability in very preterm infants born at or before 32-week gestation. DNA isolated from newborns’ dried blood spots were used for pyrosequencing to detect both SNPs. Association between EAAT2 genotypes and cerebral palsy, cystic periventricular leukomalacia and a low developmental score was then assessed. The two SNPs were concordant in 89.4% of infants resulting in three common genotypes all carrying two C and two A alleles in different combinations. However, in 10.6% of cases, non-concordance was found, generating six additional rare genotypes. The A alleles at both loci appeared to be detrimental and consequently, the risk of developing cerebral palsy increased four- and sixfold for each additional detrimental allele at -200 and -181 bp, respectively. The two SNPs altered the regulation of the EAAT2 promoter activity and glutamate homeostasis. This study highlights the significance of glutamate in the pathogenesis of preterm brain injury and subsequent development of cerebral palsy and neurodevelopmental disabilities. Furthermore, the described EAAT2 SNPs may be an early biomarker of vulnerability to neurodisability and may aid the development of targeted treatment strategies.
Similar content being viewed by others
Introduction
Progress in perinatal care over the last three decades has led to greater survival rates in infants born prematurely [1, 2]. The incidence of premature birth in developed countries varies from 7.6–12% of all births [3]. While 90% of very preterm infants (below 32-week gestation) now survive beyond the postpartum period, ~35% have neurodisabilities [4]. These disabilities include cerebral palsy, cognitive- and behavioural problems [5]. The estimated cost of preterm birth throughout childhood in England and Wales with a birth rate of 700,000/year is around £3 billion per annum [6]. Susceptibility of a preterm infant to neurodisability is difficult to predict, shows considerable variation between individuals [7] and is likely to be modulated by genetic factors [8]. Better diagnostic approaches for the early identification of infants with higher risk of neurodisability are important to facilitate the development and application of appropriate treatment strategies.
Much of the neurodisability seen in very preterm infants is caused by white matter injury, known as periventricular leukomalacia (PVL) and the subsequent disruption of normal neural connectivity [9]. While the pathogenesis of PVL remains to be established, in vitro and in vivo animal studies have identified important roles for oxidative stress, cytokine-mediated injury and glutamate-induced excitotoxicity [10, 11]. Following hypoxia-ischaemia, the excitatory neurotransmitter glutamate is released into the extracellular space, causing over-activation of ionotropic glutamate receptors present in pre-myelinating oligodendrocytes [12], which induces their excitotoxic cell death and subsequent white matter lesions [10].
In the brain, neuronal and glial excitatory amino acid transporters (EAATs) play a key role in maintaining extracellular glutamate below neurotoxic levels. The activity of the predominantly astroglial high-affinity glutamate transporter EAAT2 (also known as solute carrier family 1 member 2-SLC1A2 or the rodent ortholog glutamate transporter 1-GLT-1) is responsible for 90% of total glutamate uptake [13, 14]. Furthermore, EAAT2 has been implicated in the pathology of cerebral ischemia [15]. While ischaemic brain injury was exacerbated in transgenic mice lacking the EAAT2 protein in the brain [16], upregulation of EAAT2 provides neuroprotection [15]. EAAT2 is widely expressed in the white matter of the developing human brain [17] and upregulated in reactive astrocytes in post-mortem brain tissue of preterm infants with PVL, which may indicate a response to either hypoxic-ischemic injury or inflammation [18]. Collectively, these findings suggest that dysregulated EAAT2 activity may contribute to white matter damage.
A functional single nucleotide polymorphism (SNP) in the promoter region of the EAAT2 gene has been associated with higher serum glutamate levels in adults and consequently a worse neurological outcome after stroke [19] and also with relapsing multiple sclerosis [20]. These studies raised the intriguing possibility that similar genetic differences may enhance predisposition to neurodevelopmental impairment after preterm birth. The aim of this study was to establish the role of two closely linked functional SNPs in the EAAT2 gene promoter [19, 21] in susceptibility to brain injury and neurodisability in very preterm infants.
Materials and Methods
Patient Selection
The risk of CP in infants born <33 weeks of gestation is 30 times higher than among those born at term [22]. Therefore, our study included infants born at this vulnerable period. Newborns’ dried blood spots and clinical data were obtained from all infants born ≤32 weeks of gestation and survived to discharge in the South West of England recruited to the Avon Premature Infant Project (APIP; 1990–1993, n = 329 [23]) or received care within the neonatal unit of Gloucestershire Royal Hospital (2002–2008; n = 127); Southmead Hospital, North Bristol NHS Trust (2005–2010; n = 169) or St Michael’s Hospital, University Hospitals Bristol NHS Trust (2002–2008; n = 196). Infants with major congenital anomalies of the central nervous system and genetic syndromes that may cause neurodevelopmental impairment or cerebral palsy were excluded. The archived blood spots were fully anonymised according to the Human Tissue Act and Medical Research Council (UK) Guidance and used for research without individual informed consent as permitted by the UK newborn screening programme Code of Practice for the retention and Storage of Residual Spots (April 2005, ISBN 0955013801). From the total (n = 821 infants), 208 blood spots were not traceable, 1 was excluded with a chromosomal abnormality, ten DNA samples failed all pyrosequencing assays and 61 infants had no outcome data, leaving a total of 541 infants for the analyses (Table 1).
Sample Collection and DNA Isolation
Blood was collected from heel prick blood sampling on blood spot screening cards prepared routinely within 5–8 days of birth as part of the UK Newborn Screening Programme [http://newbornbloodspot.screening.nhs.uk]. DNA was isolated as described previously [24].
Generation of Biotinylated PCR Products for Pyrosequencing
Two sequence-specific primers (EAAT2PyroF-BIO and EAAT2PyroR; Table 2) were designed to amplify a 166 bp region of the EAAT2 promoter which included the two SNPs rs111885243:C>A or g.-200C>A (at positions -200 bp) and rs4354668:a>c or g.-181A>C (at position -181 bp) using the software provided by Qiagen Pyrosequencing. The 5′ end of the forward primer was modified with biotin. PCR reactions contained 4–6 ng of genomic DNA, 1× PCR buffer (100 mM Tris-HCl, 500 mM KCl pH 8.3), 1.5 mM MgCl2, 200 μM of each dNTP, 100 pmol of each oligonucleotide and 1 unit of high-fidelity Taq polymerase (FastStart High Fidelity Taq Polymerase, Roche Diagnostics Limited, West Sussex, UK) per reaction. Amplification was performed as follows: 95 °C for 5 min, 50 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s and final extension 72 °C for 10 min. Two additional SNPs, rs116392274 in EAAT2 and rs1835740 [21], which are involved in glutamate homeostasis, were also analysed in the cohort and data are shown as Supplementary materials.
Pyrosequencing and Sanger Sequencing
All steps were carried out as previously described (Table 2) [21, 24]. Genotypes of randomly selected samples (n = 51) from pyrosequencing were confirmed by Sanger sequencing (using ABI 3730xl 96 capillary DNA Analyzers) at Eurofins MWG Operon (Ebesberg, Germany).
Primary Astrocyte Cultures and Preparation of the EAAT2 Promoter Constructs
Primary rat astrocytes were separated from mixed glial cultures of embryonic (E20) Sprague-Dawley rat brains (Harlan, UK) using the previously described selective detachment (shaking) method [25]. Following separation at day 10 in vitro, astrocytes were maintained in T75 cell culture flasks (Corning Incorporated, New York, USA) at 37 °C in a humidified 5% CO2: 95% air atmosphere. Cells were cultured in Dulbecco’s modified Eagle’s medium (Sigma Aldrich, MO, USA) containing 4.5 g/l glucose, 29 mM sodium bicarbonate, 50 U/ml penicillin, 50 μg/ml streptomycin (Sigma Aldrich, MO, USA) and 10% (v/v) foetal bovine serum (Life Technologies Ltd., Paisley, UK). Glial fibrillary acidic protein immuno-labelling and trypan blue staining [26, 27] were used to confirm the purity and viability of the astrocyte cultures. Previously described oligonucleotides were used to amplify a 773 bp fragment of the EAAT2 promoter [19]. Genomic DNA of genotype 1 and genotype 3 was amplified in 25 μl reactions containing 2 μl genomic DNA, 1X High Fidelity PCR buffer (100 mM Tris-HCl, 500 mM KCl pH 8.3), 1.5 mM MgCl2, 200 μM of each dNTP, 100 pmol of each oligonucleotide and 1 unit of high-fidelity Taq polymerase (FastStart High Fidelity Taq Polymerase, Roche Diagnostics Limited, West Sussex, UK). Amplification was performed as follows: 1 cycle at 95 °C for 5 min, 35 cycles of 94 °C for 30 s, 65 °C for 30 s, 72 °C for 1 min and final extension at 72 °C for 10 min. Following enzyme digestion and fragment purification, the promoter fragment was inserted upstream of the firefly luciferase reporter in the pGL3-basic luciferase reporter vector.
Transfection of Astrocytes and Luciferase Reporter Gene Assay
Cells were seeded at a density of 1 × 105 per well in 1 ml of complete growth medium in a 12 well plate (Corning Incorporated, New York, USA) 24 h prior to transfection. At >80% confluency, the cells were transfected using 1 μg of EAAT2 promoter construct (EAAT2PrWT -200 bp C/C -181 bp A/A or EAAT2PrMT -200 bp A/A -181 bp C/C) and 10 μl of TransIT®-Neural Transfection Reagent (Mirus Bio, Madison, WI 5371, USA) in Opti-MEM® I Reduced Serum Media (Life Technologies Ltd., Paisley, UK). One hundred nanograms of pRL-thymidine kinase plasmid (Promega, WI, USA) containing the Renilla luciferase gene was co-transfected with each construct and used as an internal control. Forty-eight-hour post-transfection, the cells were washed and harvested for the promoter activity assay. All transfections were carried out in triplicates and all experiments were repeated three times. EAAT2 promoter activity was determined using the Dual-Luciferase Reporter (DLR) Assay System (Promega, WI, USA) following the manufacturer’s guidelines.
Patient Outcome Measures
The primary outcome measure was the diagnosis of cerebral palsy. Cerebral palsy was diagnosed when a disorder of movement and posture causing activity limitation were present at clinical examination performed at 2 years of age [28]. The secondary outcome measures were (i) cystic PVL diagnosed on a cerebral ultrasound scan during the neonatal stay and (ii) a low developmental score using standardised developmental assessment tools at 2 years of age. Cerebral ultrasound scans were performed as part of routine clinical monitoring by the clinicians in all four groups of infants. Cystic PVL was diagnosed as standard [29] (i.e. when any cystic changes were visible in the periventricular white matter on ultrasound).
Standardised developmental assessment data was available for three of the four infant groups (Table 3). The Griffith Mental Developmental Scale [30] was used for the APIP and the Gloucestershire Royal Hospital group, while the Bayley Scales of Infant Development (BSID) score (initially version II to 2006, version III after 2006 to-date) [31, 32] for the St Michael’s Hospital group. BSID-II is divided into two subscales (i) cognitive (Mental Developmental Index; MDI) and (ii) motor (Psychomotor Developmental Index; PDI). The updated BSID-III has three subscales: cognitive, language and motor PDI. Infants falling in the lowest 10th centile for either the main score (Griffith) or any of the subscales (BSID) in each infant group were defined a priori as having a low developmental score. Birth weight, gestational age at birth and physiological condition during the first 5 min after birth (Apgar scores at 1 and 5 min) were considered a priori possible confounders.
Statistical Analysis
Initially, the perinatal/intrapartum characteristics (gestation, birth weight, gender, multiple births, ethnicity and Apgar score) of the population were assessed, split by their genotype. Then, univariable associations were assessed, between the two EAAT2 genotypes and the primary and secondary outcome measures (see previous section). Due to the data coming from multiple infant groups with different developmental tools, multi-level logistic regression models were derived using the Stata 10 (Stata Corp, TX, USA) “xtlogit” command, to investigate the association of the odds of each additional polymorphic allele and the outcome measures. Adjustment for possible confounders was performed by adding the perinatal/intrapartum variables described above to the logistic regression models as continuous variables. Two sensitivity analyses were performed: (i) the analysis was repeated using single-level (rather than multi-level) modelling, and (ii) the missing covariates were imputed to allow the adjusted analysis to contain the same number of individuals as the unadjusted. Genotypes or outcome data was not imputed. Imputation was performed using multiple imputation with chained equations [33]. Details of imputation technique are available on request. All analyses were conducted with Stata 10 (Stata Corp, TX, USA) or Excel (Microsoft Corp, WA, US). All data are presented as odds ratio (OR) (95% confidence interval (CI)), mean (SD) or number (percent (%)).
Results
Simultaneous Pyrosequencing of Two SNPs in the EAAT2 Promoter
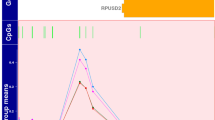
A functional SNP was reported previously in the EAAT2 promoter at -181 bp (rs4354668) [19]. Our detailed investigation of the EAAT2 promoter using Sanger sequencing revealed another SNP, 19 bp upstream of rs4354668, at position -200 bp (rs111885243) [21]. These two SNPs cannot be distinguished by single-strand conformational polymorphism used in the previous study [19]; therefore, a pyrosequencing assay was developed (Fig. 1). All traceable blood spots were analysed (n = 613) by pyrosequencing and 521 produced clear pyrograms. Ten percent of these samples (n = 51) were sequenced and the concordance with pyrosequencing was 100%. In total, 471 of the infants had clinical outcome available for the analysis of rs4354668 and rs4354668.
Pyrograms of the EAAT2 promoter SNPs. The position of the SNPs is highlighted in yellow boxes, the x-axis of each pyrogram indicates the order of reagent addition (E-enzyme, S-substrate and nucleotide A, G, T or C); the y-axis shows the light intensity generated. The numbering of pyrograms corresponds to the genotype numbers in Table 2. Due to the high GC content of the target sequence and the four C repeats before the SNP at position -181 bp, the pyrosequencing was carried out on the reverse strand. Thus, note that the sequence is in reverse orientation
Distribution of Different Alleles in the Study Population
Nine genotype combinations were identified (Table 4 and Fig. 1). In 419 samples (88.9%), the two SNPs were in linkage disequilibrium (p < 0.001). Linkage disequilibrium was not complete and hence the nine different genotypes (Table 4). The majority of alleles demonstrated high levels of concordance, such that if the -200 locus was homozygous (C/C or A/A), the -181 locus was also homozygous (A/A or C/C) and if -200 locus was heterozygous, the -181 locus was heterozygous as well (Table 4; genotypes 1–3). In the rarer genotypes (11.0% of the cases, Table 4; genotypes 4–9), the alleles were non-concordant between the two polymorphic loci. We investigated rs116392274 (g.-168C>T) in the EAAT2 promoter in the cohort but apart from one infant, who was a heterozygote, all others carried C/C alleles. Allele distribution of rs1835740 is shown in Supplementary materials.
EAAT2 Promoter Activity
To analyse the functional effects of the -200 C>A; -181A>C SNPs on transcriptional activity in vitro, genotype 1 (-200 C/C; -181 A/A) or genotype 3 (-200 A/A; -181 C/C) reporter constructs were transiently transfected into primary astrocytes, together with the pRL-TK vector as an internal control that constitutively expresses the Renilla luciferase. The genotype 1 promoter construct displayed between 4- and 4.7-fold greater activity compared with the genotype 3 construct (p < 0.0015; Fig. 2).
Promoter activity of EAAT2. Astrocytes were transiently transfected with sequences corresponding to genotype 1 (-200 C/C; -181 A/A) and 3 (-200 A/A; -181 C/C) reporter constructs. Firefly and R. reniformis luciferase activities were measured as detailed in the “Materials and Methods” section and the relative firefly/Renilla luciferase values are shown. Bars represent relative luciferase values from three independent experiments with standard deviation
Several attempts were made to measure the promoter activity of genotypes 5 and 8 using initially the three clinical samples that carried these genotypes (Table 4). These blood samples were 15–20 years old and the isolated gDNA and the resulting PCR products were of insufficient quality [24] for successful ligation to produce the required promoter constructs. In an alternative approach, we attempted to generate these variants using site directed mutagenesis of genotypes 1 and 3. However, due to the very high GC content of the promoter amplicon (over 70%; [34]), no correct mutants were obtained.
Characteristics of the Cohort
The intrapartum/perinatal characteristics of the eligible infants split by groups or genotypes are shown in Tables 1 and 5. Importantly, the patient outcome measures (e.g.: the rate of cerebral palsy (p = 0.284), cystic PVL (p = 0.553) and low developmental scores (p = 0.084)) did not differ between the four groups investigated (Table 1) and thus were combined for subsequent analysis. An association between ethnicity and genotype was observed (p < 0.001) when the whole cohort was investigated (Table 5). To better understand the nature of the association between ethnicity and the two SNPs, the cohort was investigated in more details. While there was no difference in the individual frequencies at the two SNPs by ethnicity (-181, p = 0.206 and -200, p = 0.854), white infants were more likely to show the concordance discussed above than non-white infants (94.6 vs. 76.1%, p < 0.001). Data on ethnicity was available for three of the four infant groups (Table 1) and within this population of preterm infants, there was strong evidence of deviation from the Hardy-Weinberg equilibrium (p < 0.001).
Outcome Measures
In the univariable analyses (in which associations were assessed between each of the EAAT2 SNP and the primary and secondary outcome measures independently), there was no clear evidence for an association between different alleles with cerebral palsy, cystic PVL or a low developmental score (Table 6). However, when adding both polymorphisms into the multivariable analysis, the presence of A alleles at -181 and -200 bp appeared to increase the likelihood of a low developmental score with OR of 4.56(1.53–13.60) and 3.73(1.29–10.80), respectively (Table 7; unadjusted (1)). This association persisted in the analysis adjusted for gestation, birth weight, gender and physiological condition at birth (Table 7; adjusted (2)). In contrast, there was less evidence for any association between either allele and cerebral palsy or cystic PVL. Due to the association seen with ethnicity (Table 5), this covariate was added to the model in a final adjusted analysis (Table 7; adjusted (3)). In this final model, the association with cerebral palsy strengthened with each additional A allele (locus -200 bp OR 4.34 (1.12–16.77) and locus -181 bp OR 6.64(1.76–25.07)), although there was less evidence that the polymorphism at locus -200 bp remained associated with an increased risk of a low developmental score (OR 2.84 (0.71–11.44)). Repeating the analysis using a model where the missing covariate data was imputed, the results were compatible with the main analysis. The single infant who was a heterozygote for rs116392274 had no CP or a low developmental score. Similarly, no association was observed between rs1835740 and CP or a low developmental score (Supplementary Materials).
Discussion
SNPs in EAAT2 Promoter Are Associated with Neurodevelopmental Impairment After Preterm Birth
To our knowledge, this is the first study that demonstrates association between genetic variants of EAAT2 involved in maintaining glutamate homeostasis and neurodevelopmental impairment in very preterm infants. We identified that SNP g.-200C>A in the EAAT2 promoter is strongly linked to the previously described functional SNP g.-181A>C [19], which has not been reported in earlier studies [19, 20, 35]. The A alleles at both loci appear to increase the risk of cerebral palsy and low developmental scores (Table 7). In the common concordant inheritance pattern (Table 4, genotypes 1–3), the protective C and detrimental A alleles are usually inherited together whereas in the rare non-concordant genotypes, only detrimental alleles (Tables 4 and 8, genotypes 4/5/7) or just protective alleles (Tables 4 and 8, genotypes 6/8/9) were found at both loci. This concordance was more likely with white ethnicity. Due to the strong linkage between the two SNPs, it was appropriate to enter both into the multi-level regression analysis to assess the impact of increasing detrimental A alleles. In the multi-level regression analysis (Table 7), adjustment for gestation, birth weight, gender, multiple births and Apgar scores made no significant difference to the odds of any of the outcome measures. However, the addition of ethnicity into the regression analysis strengthened the effect seen on cerebral palsy at both loci. In addition, the odds of a low developmental score were also significantly increased with each A allele at -181 bp. To put this in context, for each additional A allele at -181 or -200, the odds of cerebral palsy increased by about four- and sixfold and the odds of a low developmental score increased fourfold. The prevalence of cerebral palsy or a low developmental score was as high as 28 and 44% for genotypes 7 and 4 with three detrimental alleles, respectively (Table 8). In contrast, no association was observed between rs116392274 or rs1835740 and CP or a low development score in the cohort indicating that these SNPs are unlikely to play important roles in the injury of the developing brain (Supplementary materials).
Regulation of EAAT2 Promoter Activity
These two SNPs significantly affect EAAT2 promoter activity in vitro. The promoter fragment -742/+31 [19] containing -200A/A -181C/C sequence (genotype 3) showed a 70–80% reduction in basal EAAT2 promoter activity compared to -200C/C -181A/A (genotype 1; Fig. 2). This is a larger impact than the previously reported ~30% reduction [19]; however, in that study, the SNP at position -200 bp was not identified and it is not clear which nucleotide was present in their promoter construct. The change from A to C at -181 bp abolishes the binding site for transcription factor AP-2 (activating enhancer binding protein 2) and creates a site for GC-binding factor 2 (GCF2) which represses EAAT2 expression (Fig. 3; [19]). Reduced EAAT2 expression alters extracellular glutamate levels [13]. Despite the large difference in EAAT2 promoter activity between genotypes 3 and 1, there was no clear association with low developmental score or cerebral palsy in any of the main three genotypes (Table 8; genotypes 1–3). Similar observations were made in patients with multiple sclerosis [20] and migraine [36] where the allele and genotype frequencies for the EAAT2 promoter polymorphism were similar in patients and controls. However, the polymorphism at -181 bp was associated with higher plasma glutamate concentrations during relapsing multiple sclerosis [20].
Proposed model of the SNPs impact on EAAT2 gene regulation. a EAAT2 promoter contains a consensus binding site for transcription factor activating enhancer binding protein 2 (AP-2), which is an activator of transcription in the developing brain [53]. b Nucleotide change from A to C at -181 bp abolishes this AP-2 consensus sequence and creates a binding site for transcription factor GC-binding factor 2 (GCF2) which represses EAAT2 gene expression [19]. c, d EAAT2 promoter is not only controlled by the transcriptional machinery, but is also subject to modulation by epigenetic mechanism such as DNA methylation at CpG dinucleotides that inhibits gene expression [38, 39, 41, 42]. DNA methylation is reversible and subject to dynamic regulation throughout embryogenesis. Nucleotide changes from C to A might interfere with the normal DNA methylation process of EAAT2 at both -200 and -181 bp, affecting gene expression. The ability to downregulate EAAT2 in the developing brain seems beneficial since infants with three C alleles have better outcomes than those with only one
Gene expression in the nervous system is not only controlled by the transcriptional machinery, but it is also subject to modulation by epigenetic mechanisms such as DNA methylation [36, 37]. Dynamic DNA methylation is observed during brain development [38, 39] and the levels of DNA methylation are increased upon ischemic injury [40]. Recent studies revealed that the basal transcriptional activity of the EAAT2 gene is controlled by DNA methylation of cytosine residues in the region of -1010 to -1 bp of the EAAT2 promoter [41, 42]. Hypermethylation of the EAAT2 promoter is involved in repression of EAAT2 activation [42]. Furthermore, a recent study revealed significant differences in the methylation of ten genes involved in neuronal and glial signalling, neurotransmission, apoptosis and cellular energetics between preterm and term infants [43]. Importantly, among these genes was EAAT2, which promoter was differentially methylated at multiple CpG sites. Additionally, significant variation of EAAT2 promoter activity was observed in different brain regions and even between neighbouring cells [44]. These findings indicate that EAAT2 promoter is dynamically regulated under physiological conditions. In genotypes 4/5/7, the C alleles at both -200 and -181 bp are replaced partially or fully by A alleles (Table 8), which might interfere with the normal methylation process and the binding of GCF2 transcription factor to the EAAT2 promoter [19] (Fig. 3).
Regulation of Glutamate Level by EAAT2 in the Developing Brain
One major pathology associated with cerebral palsy is PVL [10]. Oligodendrocyte cell death is particularly prominent following hypoxia-ischemia, which leads to hypomyelination [9]. Although the causes of PVL are not completely understood, cerebral ischemia is likely to play an important role [9, 10] implicating glutamate excitotoxicity, and excessive activation of ionotropic glutamate receptors [12]. The regulation of glutamate concentration in the extracellular space by EAAT2 is therefore essential for normal synaptic function [13] as well as neuronal survival by preventing excitotoxicity [16]. However, when there is a dissipation of electrochemical gradients across the plasma membrane as occurs during hypoxia-ischemia, EAAT2 operates in reverse to release glutamate, thereby promoting excitotoxicity [45]. In a rat model, glutamate was reduced in oligodendrocytes and axons following hypoxia-ischemia suggesting that these are the main sources of glutamate in developing white matter [46]. Furthermore, EAAT2 deficient mice are more vulnerable to neuronal loss in the hippocampus following a short episode of ischemia, while the wild-type mice are more vulnerable to neuronal death following prolonged ischaemia [47]. These findings suggest that in prolonged ischaemia, EAAT2 becomes the major contributor to abnormal concentrations of extracellular glutamate. EAAT2 expression is limited primarily to oligodendrocytes early in development and is increased during the period when the premature infant is most vulnerable to PVL [17]. Furthermore, the EAAT2 protein level was found to increase substantially in some cases of PVL compared to age-related controls [18]. Similarly, a recent study showed that EAAT2 is selectively expressed in cortical layer V neurons that are damaged in premature infants with PVL [48] and hypothesised that the reversal of glutamate transport by EAAT2 together with hyperactivation of ionotropic glutamate receptors contribute to excess ambient glutamate and consequently cell death specifically in these neurons [49]. Taking together, these data indicate that in the developing white matter, it is advantageous to have the ability to dynamically downregulate EAAT2 expression during ischaemia. Our genetic data supports this hypothesis; C alleles at -200 and/or -181 bp allow for dynamic alteration of EAAT2 expression via methylation and by the binding of GCF2 transcription factor (Fig. 3). In contrast, in infants who carry mainly A alleles, regulation of EAAT2 via these mechanisms is impaired, which increases ischaemic vulnerability and subsequent impaired neurodevelopment and cerebral palsy.
Study Design Benefits and Limitations
This study included all infants of 32-week gestation or less, including multiples who survived the first 5–8 days of life. Consequently, preterm infants with severe brain injury due to hypoxia-ischaemia or intraventricular haemorrhage, who often die in the first few days of life, were not included which may explain the deviation from the Hardy-Weinberg equilibrium. Participants originated from four different infant groups/neonatal centres in the South West of England and included all ethnic groups and therefore the findings are applicable to the whole UK population of preterm infants. However, due to the retrospective design of the study, not all bloodspots could be traced from the complete population. The use of different neurodevelopmental assessment tools for the different groups precluded the use of raw cognitive or motor scores as continuous variables. The pragmatic solution was to classify those in the lowest 10th percentile of each group for each subscale/score as having a low developmental score. The lowest 10th percentile for each score translated as two standard deviations below the normal population mean, which is widely accepted as the cut-off for moderate/severe developmental impairment when using a single developmental assessment tool in clinical studies [50]. Cystic PVL was diagnosed on routine clinically directed cerebral ultrasound and white matter injury was reported and coded if it was severe and cystic in nature. We included an extra group with cystic PVL data (Table 1; Southmead Hospital) but despite increasing the power of the analysis, there was no evidence for an association between the SNPs tested and the measurable ultrasound changes. The overall proportion of cystic PVL in this work was 6.7%, which is not statistically different from the population rate in the UK Vermont Oxford dataset at the time (4.8%; p = 0.117). These data suggest that milder (non-cystic) white matter injury may not have been detected on clinical cerebral ultrasound in these groups and consequently an association with EAAT2 genotype and white matter damage was not found. Magnetic resonance imaging, which is more sensitive in detecting milder grades of white matter injury [51], is not used routinely in the UK to screen the preterm brain. These neuroimaging approaches performed in the first weeks of life are imprecise surrogate markers of neurological function. Therefore, structured functional neurological assessment at 2 years for cerebral palsy and neurodevelopmental impairment (used in this study) is considered to be the gold standard measure of neurological outcome in preterm infants [52].
Conclusions
In this study, we have found that g.-200C>A and 181A>C SNPs are associated with both clinical neurodevelopmental outcomes and measurable in vitro effects on glutamate homeostasis. These findings indicate that glutamate is likely to be involved in the pathogenesis of brain injury and subsequent development of cerebral palsy and neurodevelopmental impairments in the human infant. It is plausible that g.-200C>A SNP may also have a major effect on the development of neurological diseases in the adult population as this SNP is so closely linked to the g.-181A>C SNP, which was reported to affect neurological function after adult stroke [19], multiple sclerosis [20] and in schizophrenia [35]. The described EAAT2 SNPs may have utility as a viable early biomarker of cerebral palsy and long-term neurodisability in high-risk preterm infants. These results warrant a prospective study with complete recruitment (including non-survivors) to confirm the utility as early biomarker of neurological outcome. Our results also validate the notion that glutamate plays a pivotal role in preterm brain injury and opens the debate around exploration of glutamate uptake manipulation as potential pharmacological intervention for the prevention of preterm brain injury in infants with this genetic vulnerability. Better understanding of the dynamic transcriptional regulation of EAAT2 during the perinatal period may be key to the future development of effective clinical interventions.
References
Field DJ, Dorling JS, Manktelow BN, Draper ES (2008) Survival of extremely premature babies in a geographically defined population: prospective cohort study of 1994-9 compared with 2000-5. Br Med J 336:1221–1223. doi:10.1136/bmj.39555.670718.BE
Costeloe KL, Hennessy EM, Haider S, Stacey F, Marlow N, Draper ES (2012) Short term outcomes after extreme preterm birth in England: comparison of two birth cohorts in 1995 and 2006 (the EPICure studies). Br Med J 345:e7976. doi:10.1136/bmj.e7976
Galinsky R, Polglase GR, Hooper SB, Black MJ, Moss TJ (2013) The consequences of chorioamnionitis: preterm birth and effects on development. J Pregnancy 2013:412831. doi:10.1155/2013/412831
Moser K, Macfarlane A, Chow YH, Hilder L, Dattani N (2007) Introducing new data on gestation-specific infant mortality among babies born in 2005 in England and Wales. Health Stat Q 2007 Autumn:13–27.
Wilson-Costello D, Friedman H, Minich N, Siner B, Taylor G, Schluchter M, Hack M (2007) Improved neurodevelopmental outcomes for extremely low birth weight infants in 2000-2002. Pediatrics 119:37–45. doi:10.1542/peds.2006-1416
Mangham LJ, Petrou S, Doyle LW, Draper ES, Marlow N (2009) The cost of preterm birth throughout childhood in England and Wales. Pediatrics 123:e312–e327. doi:10.1542/peds.2008-1827
Marret S, Marchand-Martin L, Picaud J-C, Mascoët J-M, Arnaud C, Rozé JC, Truffert P, Larroque B et al, EPIPAGE Study Group (2013) Brain injury in very preterm children and neurosensory and cognitive disabilities during childhood: the EPIPAGE cohort study. PLoS One 8:e62683. doi:10.1371/journal.pone.0062683
Boardman JP, Waley A, Ball G, Takousis P, Krishnan ML, Hughes-Carre L, Aljabar P, Serag A et al (2014) Common genetic variants and risk of brain injury after preterm birth. Pediatrics 133:e1655–e1663. doi:10.1542/peds.2013-3011
Volpe JJ (2009) The encephalopathy of prematurity-brain injury and impaired brain development inextricably intertwined. Semin. Pediatr Neurol 16:167–178. doi:10.1016/j.spen.2009.09.005
Volpe JJ, Kinney HC, Jensen FE, Rosenberg PA (2011) The developing oligodendrocyte: key cellular target in brain injury in the premature infant. Int J Devl Neurosci 29:423–440. doi:10.1016/j.ijdevneu.2011.02.012
Elitt CM, Rosenberg PA (2014) The challenge of understanding cerebral white matter injury in the premature infant. Neuroscience 276:216–238. doi:10.1016/j.neuroscience.2014.04.038
Jensen FE (2005) Role of glutamate receptors in periventricular leukomalacia. J Child Neurol 20:950–959
Danbolt NC (2001) Glutamate uptake. Prog Neurobiol 65:1–105
Maragakis NJ, Rothstein JD (2004) Glutamate transporters: animal models to neurologic disease. Neurobiol Dis 15:461–473. doi:10.1016/j.nbd.2003.12.007
Chao X-D, Fei F, Fei Z (2010) The role of excitatory amino acid transporters in cerebral ischemia. Neurochem Res 35:1224–1230. doi:10.1007/s11064-010-0178-3
Tanaka K, Watase K, Manabe T, Yamada K, Watanabe M, Takahashi K, Iwama H, Nishikawa T et al (1997) Epilepsy and exacerbation of brain injury in mice lacking the glutamate transporter GLT-1. Science 276:1699–1702
DeSilva TM, Kinney HC, Borenstein NS, Trachtenberg FL, Irwin N, Volpe JJ, Rosenberg PA (2007) The glutamate transporter EAAT2 is transiently expressed in developing human cerebral white matter. J Comp Neurol 501:879–890. doi:10.1002/cne.21289
DeSilva TM, Billiards SS, Borenstein NS, Trachtenberg FL, Volpe JJ, Kinney HC, Rosenberg PA (2008) Glutamate transporter EAAT2 expression is up-regulated in reactive astrocytes in human periventricular leukomalacia. J Comp Neurol 508:238–248. doi:10.1002/cne.21667
Mallolas J, Hurtado O, Castellanos M, Blanco M, Sobrino T, Serena J, Vivancos J, Castillo J et al (2006) A polymorphism in the EAAT2 promoter is associated with higher glutamate concentrations and higher frequency of progressing stroke. J Exp Med 203:711–717. doi:10.1084/jem.20051979
Pampliega O, Domercq M, Villoslada P, Sepulcre J, Rodríguez-Antigüedad A, Matute C (2008) Association of an EAAT2 polymorphism with higher glutamate concentration in relapsing multiple sclerosis. J Neuroimmunol 195:194–198. doi:10.1016/j.jneuroim.2008.01.011
Rajatileka S, Luyt K, Williams M, Harding D, Odd D, Molnár E, Váradi A (2014) Detection of three closely located single nucleotide polymorphisms in the EAAT2 promoter: comparison of single-strand conformational polymorphism (SSCP), pyrosequencing and Sanger sequencing. BMC Genet 15:80. doi:10.1186/1471-2156-15-80
MacLennan AH, Thompson SC, Gecz J (2015) Cerebral palsy: causes, pathways, and the role of genetic variants. Am J Obstet Gynaecol 213:779–788. doi:10.1016/j.ajog.2015.05.034
Avon Premature Infant Project (1998) Randomised trial of parental support for families with very preterm children. Arch Dis Child Fetal Neonatal Ed 79:F4–11. doi:10.1136/fn.79.1.F4
Rajatileka S, Luyt K, El-Bokle M, Williams M, Kemp H, Molnár E, Váradi A (2013) Isolation of human genomic DNA for genetic analysis from premature neonates: a comparison between newborn dried blood spots, whole blood and umbilical cord tissue. BMC Genet 14:105. doi:10.1186/1471-2156-14-105
McCarthy KD, de Vellis J (1980) Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J Cell Biol 85:890–902
Luyt K, Varadi A, Molnar E (2003) Functional metabotropic glutamate receptors are expressed in oligodendrocyte progenitor cells. J Neurochem 84:1452–1464. doi:10.1046/j.1471-4159.2003.01661.x
Luyt K, Slade TP, Dorward JJ, Durant CF, Wu Y, Shigemoto R, Mundell SJ, Váradi A et al (2007) Developing oligodendrocytes express functional GABAB receptors that stimulate cell proliferation and migration. J Neurochem 100:822–840. doi:10.1111/j.1471-4159.2006.04255.x
Bax M, Goldstein M, Rosenbaum P, Leviton A, Paneth N, Dan B, Jacobsson B, Damiano D et al, Executive Committee for the Definition of Cerebral Palsy (2005) Proposed definition and classification of cerebral palsy, April 2005. Dev Med Child Neurol 47:571–576
de Vries LS, Eken P, Dubowitz LM (1992) The spectrum of leukomalacia using cranial ultrasound. Behav Brain Res 49:1–6
Huntley M (1996) The Griffiths Mental Development Scales: from birth to 2 years. Association for Research in Infant and Child Development (ARICD). Oxford, UK: The Test Agency 1996:5–39.
Bayley N (1993) Manual for the Bayley scales of infant development, 2nd edition edn. Psychological Corporation, San Antonio, Texas
Bayley N (2006) Bayley scales of infant development, 3rd edition edn. Harcourt Assessment, San Antonio, Texas
Odd DE, Lewis G, Whitelaw A, Gunnell D (2009) Resuscitation at birth and cognition at 8 years of age: a cohort study. Lancet 373:1615–1622. doi:10.1016/S0140-6736(09)60244-0
Sun D, Ostermaier MK, Heydenreich FM, Mayer D, Jaussi R, Standfuss J, Veprintsev DB (2013) AAscan, PCRdesign and MutantChecker: a suite of programs for primer design and sequence analysis for high-throughput scanning mutagenesis. PLoS One 8:e78878. doi:10.1371/journal.pone.0078878
Spangaro M, Bosia M, Zanoletti A, Bechi M, Cocchi F, Pirovano A, Lorenzi C, Bramanti P et al (2012) Cognitive dysfunction and glutamate reuptake: effect of EAAT2 polymorphism in schizophrenia. Neurosci Lett 522:151–155. doi:10.1016/j.neulet.2012.06.030
Shin HE, Han SJ, Lee KS, Park JW (2011) Polymorphism of the glutamate transporter protein EAAT2 and migraine transformation into chronic daily headache. J Clin Neurol 7:143–147. doi:10.3988/jcn.2011.7.3.143
Feng J, Fouse S, Fan G (2007) Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res 61:58R–63R. doi:10.1203/pdr.0b013e3180457635
Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y et al (2013) Global epigenomic reconfiguration during mammalian brain development. Science 341:1237905. doi:10.1126/science.1237905
Spiers H, Hannon E, Schalkwyk LC, Smith R, Wong CC, O'Donovan MC, Bray NJ, Mill J (2015) Methylomic trajectories across human fetal brain development. Genome Res 25:338–352. doi:10.1101/gr.180273.114
Endres M, Meisel A, Biniszkiewicz D, Namura S, Prass K, Ruscher K, Lipski A, Jaenisch R et al (2000) DNA methyltransferase contributes to delayed ischemic brain injury. J Neurosci 20:3175–3181
Zschocke J, Allritz C, Engele J, Rein T (2007) DNA methylation dependent silencing of the human glutamate transporter EAAT2 gene in glial cells. Glia 55:663–674. doi:10.1002/glia.20497
Yang Y, Gozen O, Vidensky S, Robinson MB, Rothstein JD (2010) Epigenetic regulation of neuron-dependent induction of astroglial synaptic protein GLT1. Glia 58:277–286. doi:10.1002/glia.20922
Sparrow S, Manning JR, Cartier J, Anblagan D, Bastin ME, Piyasena C, Pataky R, Moore EJ et al (2016) Epigenomic profiling of preterm infants reveals DNA methylation differences at sites associated with neural function. Transl Psychiatry 6:e716. doi:10.1038/tp.2015.210
de Vivo L, Melone M, Rothstein JD, Conti F (2010) GLT-1 promoter activity in astrocytes and neurons of mouse hippocampus and somatic sensory cortex. Front Neuroanat 3:31. doi:10.3389/neuro.05.031.2009
Fern R, Möller T (2000) Rapid ischemic cell death in immature oligodendrocytes: a fatal glutamate release feedback loop. J Neurosci 20:34–42
Back SA, Craig A, Kayton RJ, Luo NL, Meshul CK, Allcock N, Fern R (2007) Hypoxia-ischemia preferentially triggers glutamate depletion from oligodendroglia and axons in perinatal cerebral white matter. J Cereb Blood Flow Metab 27:334–347. doi:10.1038/sj.jcbfm.9600344
Mitani A, Tanaka K (2003) Functional changes of glial glutamate transporter GLT-1 during ischemia: an in vivo study in the hippocampal CA1 of normal mice and mutant mice lacking GLT-1. J Neurosci 23:7176–7182
Andiman SE, Haynes RL, Trachtenberg FL, Billiards SS, Folkerth RD, Volpe JJ, Kinney HC (2010) The cerebral cortex overlying periventricular leukomalacia: analysis of pyramidal neurons. Brain Pathol 20:803–814. doi:10.1111/j.1750-3639.2010.00380.x
DeSilva TM, Borenstein NS, Volpe JJ, Kinney HC, Rosenberg PA (2012) Expression of EAAT2 in neurons and protoplasmic astrocytes during human cortical development. J Comp Neurol 520:3912–3932. doi:10.1002/cne.23130
Wood NS, Marlow N, Costeloe K, Gibson AT, Wilkinson AR (2000) Neurologic and developmental disability after extremely preterm birth. EPICure study group. N Engl J Med 343:378–384. doi:10.1056/NEJM200008103430601
Miller SP, Cozzio CC, Goldstein RB, Ferriero DM, Partridge JC, Vigneron DB, Barkovich AJ (2003) Comparing the diagnosis of white matter injury in premature newborns with serial MR imaging and transfontanel ultrasonography findings. AJNR Am J Neuroradiol 24:1661–1669
Marlow N (2004) Neurocognitive outcome after very preterm birth. Arch Dis Child Fetal Neonatal Ed 89:F224–F228
Mitchell PJ, Timmons PM, Hébert JM, Rigby PW, Tjian R (1991) Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev 5:105–119
Acknowledgements
We would like to thank Dr. Helena Kemp for assisting with tracing the dried blood spots from the screening archive, Professor Neil Marlow for providing data on the Avon Premature Infant Project (APIP) infants, Dr. Sam O’Hare and Dr. Manal el Bokle for performing data collection on the Southmead Hospital infants and Dr. Sally Jary for performing neurodevelopmental assessments on the St Michael’s Hospital infants.
Author Contributions
SR: study design, sample preparation for experiments, pyrosequencing, data interpretation, manuscript preparation. DO: patient data collection and analysis, statistical analysis, preparation of manuscript. MTR: Promoter construct generation. ACS and LD: primary astrocyte generation and transfection for promoter assay. MW and CC: pyrosequencing assay optimisation. DH, MW and MO: clinical sample and data collection. JC: manuscript preparation. EM, KL and AV: conception, design of the study, manuscript preparation, supervision of the experimental work, funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The study received ethical approval in April 2010 from the National Research Ethics Service, UK (REC reference number 10/H0106/10).
Conflict of Interest
EM is member of the Scientific Advisory Board of Hello Bio [www.hellobio.com].
Funding
This work was supported by the University of the West of England, Bristol, UK (AV). EM is supported by the Biotechnology and Biological Sciences Research Council, UK (grants BB/F011326/1 and BB/J015938/1). The blood spot retrieval was funded by the David Telling Charitable Trust (KL).
Additional information
Shavanthi Rajatileka and David Odd are joint first authors.
EM, KL and AV contributed equally to this paper.
Electronic Supplementary Material
Esm 1
(DOCX 64 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Rajatileka, S., Odd, D., Robinson, M.T. et al. Variants of the EAAT2 Glutamate Transporter Gene Promoter Are Associated with Cerebral Palsy in Preterm Infants. Mol Neurobiol 55, 2013–2024 (2018). https://doi.org/10.1007/s12035-017-0462-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-017-0462-1