Abstract
RNA viruses cause most of the dangerous communicable diseases. Due to their high mutation rates, RNA viruses quickly evade selective pressures and can adapt to a new host. Therefore, new antiviral approaches are urgently needed, which target more than one specific virus variant and which would optimally prevent development of viral resistance. Among the family of coronaviruses (CoV), several human pathogenic strains (HCoV) are known to cause respiratory diseases and are implied in enteric diseases. While most strains contribute to common cold-like illnesses, others lead to severe infections. One of these viruses is the newly emerged (2012), highly pathogenic Middle East respiratory syndrome coronavirus (MERS-CoV) of zoonotic origin. MERS-CoV causes a severe respiratory infection with a high mortality rate of 35 %. There is no specific treatment or infection prevention available. Here, we show that the bacterial ribonuclease Binase is able to inhibit the replication of MERS-CoV and of the low-pathogenic human coronavirus 229E (HCoV-229E) in cell culture. We demonstrate that at non-toxic concentrations, Binase decreased the titers of MERS-CoV and HCoV-229E. On a molecular level, Binase treatment reduced (i) the viral subgenomic RNAs and (ii) the viral nucleocapsidprotein (N) and non-structural protein 13 (nsp13) accumulation. Furthermore, we show that the quantity of the replication/transcription complexes within the infected cells is diminished. Thus, the data obtained might allow further development of new anti-coronaviral approaches affecting viral replication, independent of the specific virus strain.
Similar content being viewed by others
1 Introduction
The extracellular guanyl-preferring ribonuclease (RNase) Binase from non-pathogenic bacteria Bacillus pumilus is a well-studied enzyme, which has shown various biological activities in cell cultures [1]. Binase is a thermostable enzyme and does not require any co-factors or metal ions for its enzymatic action. Binase is able to enter into the eukaryotic cell cytoplasm and nucleus in 1 and 3 hours after its addition to cell culture media, respectively, and no intracellular Binase degradation by protease was detected during 48 h [2]. Media concentrations of Binase below 300 μg/ml are non-toxic for a variety of tested cell lines [2, 3]. The enzyme lacks superantigen properties to induce the polyclonal T-cell immune response [4]. Antiviral activity of Binase was demonstrated in animal models against negative-stranded RNA viruses, such as rabies virus and influenza A and B viruses [5–7]. Previously, we have reported antiviral properties of Binase against pandemic influenza A (H1N1) virus in human cell culture [3, 7]. Also, antiviral action of Binase against the positive stranded foot-and-mouth-disease virus (FMDV) has been reported. It was demonstrated that the effect of Binase on the viral RNA of FMDV was three times higher than its effect on cellular RNA [5].
MERS-CoV and HCoV-229 are common viruses causing respiratory illnesses. Both are targeted using different antiviral tools. However, no antiviral agents are absolutely successful against all types of corona viruses, especially against MERS-CoV. As a result, several outbreaks occurred which led to a massive loss of human lives in different Middle East countries and South Korea during the last years [8]. Still, there is no vaccine or specific treatment currently available to prevent MERS-CoV infection. In this case, the well-known antiviral agent Binase can provide a new antiviral strategy against corona viruses. Based on the previously reported antiviral properties of Binase, we analyzed the effect of Binase on the replication of the alpha-coronavirus HCoV-229E and the beta-coronavirus MERS-CoV in hepatoma cell line Huh7 and primary MRC5. HCoV-229E causes self-limiting upper respiratory tract infections such as the common cold, while MERS-CoV is associated with high case fatality rates and is considered by the global health community to be a potential pandemic agent, since person-to-person transmission can occur and effective therapeutic options are limited [8]. Our data provides further insight into the mode of action exerted by Binase against positive-stranded RNA viruses and could support the development of novel antiviral approaches against CoV including MERS-CoV.
2 Material and Methods
2.1 Bacteria Growth Condition and Binase Isolation
Binase (EC 3.1.27.3; single chain of 109 amino acids, molecular weight of 12.2 kDa) was collected from the culture fluid of Gram-positive bacteria Bacillus pumilus B3073 (the former name Bacillus intermedius B3073 [9], Russian National Collection of Industrial Microorganisms). Bacteria were grown in the complex phosphate deficient medium containing 2.0 % low phosphate peptone, 1.0 % glucose, 1.0 % CaCl2, 0.03 % MgSO4 × 7H2O, 0.3 % NaCl, 0.01 % MnSO4, pH 8.5 at 37 °C until the end of bacterial exponential phase. After pelleting bacteria cells using a benchtop microcentrifuge at 6000×g for 30 min, the supernatant was acidified by glacial acetic acid to pH 5.0, diluted five times with deionized water and applied on the column with DE-32 cellulose (Whatman, UK), and equilibrated with 10 mM Na acetate buffer (pH 5.0) to escape the negatively charged enzyme. The flow through from DE-32 was transferred to column packed with phosphocellulose P11 (Whatman, UK) and equilibrated with 10 mM Na acetate buffer (pH 5.0). Then, the column was equilibrated with 20 mM Na phosphate buffer (pH 7.0). Binase was eluted with 200 mM Na phosphate buffer (pH 7.0). Further polishing purification of Binase was performed using the MonoS HR 10/10 column (Sigma-Aldrich, USA) equilibrated with 20 mM Na acetate buffer (pH 5.0) and then eluted using a linear gradient of 0–0.25 M NaCl. The protein samples were lyophilized using the FreeZone 2.5 Liter Freeze Dry System (Labconco, USA) according to the manufacturer’s instructions. Homogeneity of the purified Binase was checked using the PAAG electrophoresis, Western blotting, and MALDI TOF/TOF mass spectrometry on UltrafleXtreme (Bruker Corporation, Germany) as described in [10].
2.2 Enzymatic Activity of Binase
The enzymatic activity of Binase was conducted by 260 nm absorbance measuring of the products of Binase hydrolysis of high molecular weight RNA from yeast. Binase was incubated in 0.25 M Tris–HCl buffer (pH 8.5) in the presence of 0.5 μg/μl RNA (final concentration) at 37 °C for 15 min. The reaction was stopped by addition of ice cold 6.8 % perchloric acid followed by incubation on ice bath for 10 min. Non-degraded RNA was pelleted by centrifugation at 12,000×g for 10 min, and the supernatant was used for the 260 nm absorbance measurement using the SmartSpec Plus Spectrophotometer (Bio-rad, USA). One unit of ribonuclease activity corresponded to the quantity of enzyme that increased the extinction of acid-soluble products of RNA hydrolysis at 260 nm by 1 optical unit after incubation at 37 °C for 1 h, calculated per ml of the enzyme solution. One milligram per milliliter of purified Binase had catalytic activity of 1.0–1.2 units/ml.
2.3 Cells and Viruses
Huh7 cells (hepatocellular carcinoma) and MRC5 cells (human fetal lung fibroblast) were cultured in DMEM (Gibco, USA) media supplemented with 10 % fetal calf serum (FCS), 100 U/ml penicillin and 100 μg/ml streptomycin at 37 °C, and 5 % CO2. Middle East respiratory syndrome coronavirus (MERS-CoV) and Human coronavirus (HCoV-229E) viruses were provided by the strain collection of the Institute of Medical Virology, University of Giessen, Germany, and where propagated either at 37 and 33 °C, respectively.
2.4 Cell Viability
To determine the cytotoxic concentration, which reduces cell viability by 50 % (CC50), Binase was added to culture medium of Huh7 cells (33 °C) and MRC5 cells (33 and 37 °C) at the indicated concentrations for 24 h. The cell viability after Binase incubation was determined via MTT assay [11]. CC50 was calculated using the GraphPad Prism 5.0 Software (GraphPad Software Inc., USA) by plotting the percentage of viable cells as a function of the compound concentration.
2.5 Effective Concentration
The effective concentration, which reduces the virus titer by 50 % (EC50), was determined by incubation of the cells infected with a multiplicity of infection (MOI) of 0.5 with Binase for 24 h at the indicated concentrations and subsequent virus titer analysis via focus assay using mouse anti-HCoV-229E nucleocapsid protein monoclonal antibody (Ingenasa, Spain) [12]. EC50 was calculated using the GraphPad Prism 5.0 Software (GraphPad Software Inc., USA) by plotting the percentage of virus titers as a function of the compound concentration.
2.6 Western Blot Analysis
Western Blot analysis was used to detect the viral N and nsp13 proteins in HCoV-229E-infected Huh7 cells at 33 °C with and without Binase treatment using mouse anti-HCoV-229E nucleocapsidprotein monoclonal antibody (Ingenasa, Spain), polyclonal rabbit anti-nsp13 serum (Institute for Medical Virology, Justus Liebig University, Giessen, Germany), rabbit anti-beta-actin monoclonal antibody (Abcam, USA), and mouse anti-beta-actin monoclonal antibody (Abcam, USA). The quantitative analysis of Western Blot results was performed using the Quantity One software (Bio-rad, USA).
2.7 Northern Blot Analysis
Northern Blot analysis was used to analyze the effect of Binase on the intra-cellular amount of coronaviral mRNAs, which are formed during viral replication of Huh/cells infected with HCoV-229E at 33 °C [13]. 5S RNA was not used as a loading control because its position interfered with the coronaviral mRNAs. Therefore, the same amount (5 μg) of cellular RNAs from non-infected, infected with or without Binase treatment was used. The quantitative analysis was performed using the Quantity One software (Bio-rad, USA).
2.8 Immunofluorescence Analysis
The Huh7 cells were infected with HCoV-229E at 3 MOI and proceeded with or without Binase treatment. To detect the viral dsRNA and nsp8 protein, the cells were fixed after 24 h p.i. using ice-cold methanol and stained with mouse anti-dsRNA mAb (1:100, J2, English & Scientific Consulting Kft, Hungary) and polyclonal rabbit anti-HCoV-229E nsp8 serum (1:100). Antibodies/sera were diluted in PBS containing 3 % BSA. The goat anti-mouse IgG Alexa Fluor 594 and Alexa Fluor 488-labeled goat anti-rabbit IgG F(ab’) fragment (1:500, Invitrogen, USA) were used as secondary antibodies for detection using a laser-scanning confocal microscope (Leica SP05 CLSM, Germany). The dsRNA and nsp8 production of CoV-229E in Binase-treated and virus-infected Huh7 cells were quantified using the Leica Application Suite X (Leica Microsystems, Germany) and open-source software ImageJ. Analysis was performed in three biological replicates.
2.9 Biosafety
All experiments involving coronaviruses were performed using biosafety level 2 (BSL2) or biosafety level 3 (BSL3) containment laboratory approved for such use by the local authorities (RP, Giessen, Germany).
3 Results and Discussion
3.1 Binase is Well Tolerated by Huh7 and MRC5 Cells
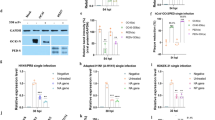
Previously, it was shown that at a concentration of 490 μg/ml, Binase is non-toxic towards human A549 cells [3]. Here, we show that Binase did not exert a cytotoxic effect towards Huh7 at 33 °C (Fig. 1a) and primary MRC5 cells at 33 and 37 °C (Fig. 1b, c) up to a concentration of 500 μg/ml at 24-h incubation time. These results show that the toxic effect of Binase is comparable for different cell types.
Cell viability and virus inhibition. Huh7 cells incubated at 33 °C (a) and MRC5 cells incubated at 33 °C (b) and at 37 °C (c) were treated with Binase at the indicated concentrations for 24 h. Cell viability (%) was determined via MTT assay (a–c). Huh7 cells and MRC5 cells infected with HCoV-229E at 33 °C (d, e), as well as MRC5 cells infected with MERS-CoV at 37 °C (f) were treated with Binase at the indicated concentrations. At 24 h p.i., the virus titer (%) was determined via focus forming assay and EC50 was calculated
3.2 Replication of HCoV-229E and MERS-CoV in Different Cell Lines is Efficiently Reduced by Binase Treatment
The effective concentration (EC50) of Binase against HCoV-229E determined 24 h post infection (p.i.) was found to be 0.4 μg/ml in infected Huh7 cells (33 °C) and 43 μg/ml in infected MRC5 cells at 33 °C (Fig. 1d, e). Therefore, Binase was 107-fold more effective against HCoV-229E in Huh7 cells than in primary MRC5 cells. As the EC50 of Binase against MERS-CoV was 2 μg/ml in infected MRC5 cells (37 °C), Binase was found to be 22-fold more effective against MERS-CoV than against HCoV-229E in the MRC5 cell culture (Fig. 1e, f). Concerning the fact that Binase was non-toxic up to 500 μg/ml, the selectivity index (SI: CC50/EC50) is at least 1250, 11,6, and 250 for HCoV-229E-infected Huh7 cells (33 °C), HCoV-229E-infected MRC5 cells (33 °C), and MERS-CoV-infected MRC5 cells (37 °C), respectively.
3.3 Binase Treatment of Infected Cells Reduces Viral RNA and Protein Levels
The coronaviral subgenomic RNAs 2, 4, 5, 6, and 7 were not detected in the HCoV-229E virus-infected Huh7 cells after their treatment with Binase at 1 and 10 μg/ml except for RNA 7 which was slightly visible when 1 μg/ml Binase was used (Fig. 2a). The quantitative analysis of the Nothern blot results showed the decrease of all corona viral subgenomic mRNAs by 86.0 ± 0.8 % and 97 ± 1.3 % upon 1 and 10 μg/ml Binase treatment, respectively, as compared to the subgenomic mRNAs level in virus-infected Binase-non-treated cells which was taken as 100 %. The corona viral structural N and non-structural nsp13 protein levels in the cytoplasm of HCoV-229E-infected Huh7 cells were analyzed by Western Blot (Fig. 2b). It was detected that Binase reduced the level of viral N protein by 43.3 ± 5.5 % (10 μg/ml Binase) and 1.5 ± 1.1 % (1 μg/ml Binase). It also decreased the level of nsp13 protein by 51.5 ± 3.4 % (10 μg/ml Binase) and 21.4 ± 3.1 % (1 μg/ml Binase). The amount of the cellular housekeeping protein actin was not affected by the Binase concentration to a similar extent. These results indicate that incubation of CoV-infected cells with media containing Binase at non-toxic concentration leads to a strong reduction of intra-cellular viral subgenomic RNAs, which is likely to contribute to the reduction in the amount of viral proteins. The finding that this reduction of the different subgenomic RNAs is much stronger than the reduction of the viral proteins might be due to the fact that the effect of the Binase treatment was determined 24 h p.i.. As several proteins can be translated from one mRNA, it can be imagined that a certain amount of viral proteins to be produced before the less stable subgenomic RNAs is reduced to the extent as detected after 24 h p.i.. The more stable proteins remain and can be therefore detected at the time point of investigation. The prominent effect of the Binase treatment on the viral RNAs in the HCoV-229E-infected Huh7 cells, including the full length genomic RNA, coincides with a strong reduction in the virus titer.
Binase affects coronaviral RNA and protein production. Northern blot hybridization demonstrating the effect of Binase on the HCoV-229E subgenomic mRNAs accumulation in the virus-infected Huh7 cells after 24 h p.i. (a). Western blot analysis of HCoV-229E viral structural protein N and non-structural protein nsp13 levels in virus-infected Huh7 cells after 24 h p.i. using actin, a product of housekeeping gene of the cells as loading control (b). Immunofluorescence confocal microscopy of Binase-treated HCoV-229E-infected Huh7 cells at 24 h p.i.(c). dsRNA (red) and the nsp8 protein (green) were used as markers of the CoV replication and transcription complexes. The nuclear DNA (blue) was stained with DAPI. Scale, 50 μm
3.4 The Quantity of Viral Replication and Transcription Complexes is Decreased after Binase Treatment of Infected Cells
Confocal microscopy revealed a stronger Binase-dependent decrease in the amount of viral intracellular markers, dsRNA, and nsp8, which represent coronaviral replication and transcription complex formation inside the infected cells at Binase concentration of 10 than at 1 μg/ml in the media (Fig. 2c). dsRNA and nsp8 were reduced by 49.4 ± 3.0 % and 87.3 ± 1.7 %, respectively, upon 10 μg/ml Binase treatment, while at 1 μg/ml Binase, their levels were decreased by 17.0 ± 7.3 % and 54.7 ± 4.4 %, respectively. The reduction of RTCs is in line with the data obtained from the Western blot analysis and is reflected by the decreased virus titers. Nevertheless, the effect of Binase treatment on the subgenomic RNAs (Fig. 2a) is much stronger than that on the dsRNA and nsp8 detected by immunofluorescence. Notably, the RTCs are anchored inside double-membrane vesicles (DMVs) [14, 15]. Therefore, it is tempting to speculate that in contrast to the subgenomic RNAs, which were also found in the cytoplasm [16], the dsRNA is partially protected from the effect of the Binase treatment.
4 Conclusions
The results obtained so far indicate that Binase in the cell culture media is non-toxic for Huh7 and MRC5 cells up to a concentration of 500 μg/ml. In coronavirus-infected cells, Binase treatment impairs the production of viral RNAs and consequently of viral proteins at concentrations well blow the highest non-toxic concentration, resulting in decreased virus titers. The anti-alpha- and anti-betacoronavirus activities of Binase indicate a broad anti-coronaviral effect. These findings could be useful to develop novel antiviral approaches against different strains of coronaviruses.
References
Ulyanova, V., Vershinina, V., Ilinskaya, O. (2011). Barnase and binase: twins with distinct fates. FEBS Journal, 278(19), 3633–3643. doi:10.1111/j.1742-4658.2011.08294.x.
Cabrera-Fuentes, H. A., Aslam, M., Saffarzadeh, M., Kolpakov, A., Zelenikhin, P., Preissner, K. T., et al. (2013). Internalization of Bacillus intermedius ribonuclease (BINASE) induces human alveolar adenocarcinoma cell death. Toxicon, 69, 219–226. doi:10.1016/j.toxicon.2013.03.015.
Shah Mahmud, R., & Ilinskaya, O. N. (2013). Antiviral activity of binase against the pandemic influenza a (H1N1) virus. Acta Naturae, 5(4), 44–51.
Ilinskaya, O. N., Zelenikhin, P. V., Petrushanko, I. Y., Mitkevich, V. A., Prassolov, V. S., Makarov, A. A. (2007). Binase induces apoptosis of transformed myeloid cells and does not induce T-cell immune response. Biochemical and Biophysical Research Communications, 361(4), 1000–1005. doi:10.1016/j.bbrc.2007.07.143.
Alekseeva, I. I., Kurinenko, B. M., Kleiner, G. I., Skuia, A., Penzikova, G. A. (1981). Comparative study of the antiviral activity of pancreatic and microbial RNAse. Antibiotiki, 26(7), 527–532.
Shneider, M. A., Shtil’bans, E. B., Kuprianov-Ashin, E. G., Potselueva, L. A., Zaikonnikova, I. V., Kurinenko, B. M. (1990). Anti-influenza effect of bacterial RNAse and the pharmacokinetic basis of its administration in experimental studies. Antibiotiki i Khimioterapiya, 35(3), 27–31.
Ilinskaya, O. N., & Shah Mahmud, R. (2014). Ribonucleases as antiviral agents. Molecular Biology, 48(5), 615–623.
Chan, J. F., Lau, S. K., To, K. K., Cheng, V. C., Woo, P. C., Yuen, K. Y. (2015). Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clinical Microbiology Reviews, 28(2), 465–522. doi:10.1128/CMR.00102-14.
Ulyanova, V., Shah Mahmud, R., Dudkina, E., Vershinina, V., Domann, E., Ilinskaya, O. (2016). Phylogenetic distribution of extracellular guanyl-preferring ribonucleases renews taxonomic status of two Bacillus strains. Journal of General and Applied Microbiology, 62(4), 181–188. doi:10.2323/jgam.2016.02.005.
Dudkina, E., Ulyanova, V., Shah Mahmud, R., Khodzhaeva, V., Dao, L., Vershinina, V., et al. (2016). Three-step procedure for preparation of pure Bacillus altitudinis ribonuclease. FEBS Open Bio, 6(1), 24–32. doi:10.1002/2211-5463.12023.
Mostafa, A., Kanrai, P., Petersen, H., Ibrahim, S., Rautenschlein, S., Pleschka, S. (2015). Efficient generation of recombinant influenza a viruses employing a new approach to overcome the genetic instability of HA segments. PloS One, 10(1), e0116917. doi:10.1371/journal.pone.0116917.
Matrosovich, M., Matrosovich, T., Garten, W., Klenk, H. D. (2006). New low-viscosity overlay medium for viral plaque assays. Virology Journal, 3, 63. doi:10.1186/1743-422X-3-63.
Thiel, V., Ivanov, K. A., Putics, A., Hertzig, T., Schelle, B., Bayer, S., et al. (2003). Mechanisms and enzymes involved in SARS coronavirus genome expression. Journal of General Virology, 84(Pt 9), 2305–2315. doi:10.1099/vir.0.19424-0.
Hagemeijer, M. C., Verheije, M. H., Ulasli, M., Shaltiel, I. A., de Vries, L. A., Reggiori, F., et al. (2010). Dynamics of coronavirus replication-transcription complexes. Journal of Virology, 84(4), 2134–2149. doi:10.1128/JVI.01716-09.
Oostra, M., te Lintelo, E. G., Deijs, M., Verheije, M. H., Rottier, P. J., de Haan, C. A. (2007). Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. Journal of Virology, 81(22), 12323–12336. doi:10.1128/JVI.01506-07.
Fung, T. S., & Liu, D. X. (2014). Coronavirus infection, ER stress, apoptosis and innate immunity. Frontiers in Microbiology, 5, 296. doi:10.3389/fmicb.2014.00296.
Acknowledgments
The work was supported by the KFU, Russia, and JLU Giessen, Germany, partnership program and the program for competitive growth of KFU. This work was also supported in part by grants from the Russian Science Foundation (grant no. 14-14-00522), the research visits program “Research stays for university academics and scientists” by the German Academic Exchange Service (DAAD), the German Ministry of Education and Research (BMBF)-funded German Centre for Infection Research (DZIF), partner site Giessen, Germany (TTU Emerging Infections), and the German Research Foundation (DFG)-funded Collaborative Research Centre SFB 1021 (RNA viruses: RNA metabolism, host response and pathogenesis, TP C1).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Müller, C., Ulyanova, V., Ilinskaya, O. et al. A Novel Antiviral Strategy against MERS-CoV and HCoV-229E Using Binase to Target Viral Genome Replication. BioNanoSci. 7, 294–299 (2017). https://doi.org/10.1007/s12668-016-0341-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12668-016-0341-7