Abstract
Cancer with heavily economic and social burden is the hot point in the field of medical research. Some remarkable achievements have been made; however, the exact mechanisms of tumor initiation and development remain unclear. Cancer is a complex, whole-body disease that involves multiple abnormalities in the levels of DNA, RNA, protein, metabolite and medical imaging. Biological omics including genomics, transcriptomics, proteomics, metabolomics and radiomics aims to systematically understand carcinogenesis in different biological levels, which is driving the shift of cancer research paradigm from single parameter model to multi-parameter systematical model. The rapid development of various omics technologies is driving one to conveniently get multi-omics data, which accelerates predictive, preventive and personalized medicine (PPPM) practice allowing prediction of response with substantially increased accuracy, stratification of particular patients and eventual personalization of medicine. This review article describes the methodology, advances, and clinically relevant outcomes of different “omics” technologies in cancer research, and especially emphasizes the importance and scientific merit of integrating multi-omics in cancer research and clinically relevant outcomes.
Similar content being viewed by others
Introduction
The high-mortality cancer [1] experiences a process of complex and multistep development, malignant cells acquired eight biological capabilities, including sustaining proliferative signaling, evading growth suppressors, resisting cell death, inducing angiogenesis, activating invasion and metastasis, enabling replicative immortality, reprogramming of energy metabolism and evading immune destruction, which are regarded as the hallmarks of cancer [2]. Despite remarkable achievements in cancer research, the exact mechanism of tumor initiation and development still remain unclear yet. Since the Human Genome Project, the emerging scientific era of “omics” has revolutionized the study of cancer [3] (Fig. 1). Omics technologies are primarily aimed at the comprehensive detection of genes (genomics), RNAs (transcriptomics), proteins (proteomics), metabolites (metabolomics), and quantitative features of medical imaging (radiomics) [4]. Omics technologies have a wide-range application in both basic research and clinical treatment of cancer. Based on the next-generation sequencing (NGS), genomics and transcriptomics provide one with a better understanding of the structure of cancer genome and discover differentially expressed genes that drive and maintain tumorigenesis [5,6,7,8,9,10,11]. More importantly, this genome profiling has the potential role in establishing different molecular subtypes and stratification of different patients, which is crucial in precisely personalized treatment. High performance liquid-chromatography (HPLC), mass spectrometry (MS), and nuclear magnetic resonance (NMR) technologies are widely used in discovery of new biomarkers and drug targets from cancer proteome and metabolome [12,13,14,15,16,17,18]. These biomarkers, including predictive biomarkers for treatment stratification, diagnostic biomarkers for early detection, and prognostic biomarkers for estimation of patient clinical outcome, are important for the predicition and prevention of tumors. At the same time, some key molecules in the pathway and network of tumors such as proteins and metabolites can be recognized as targets for targeted therapy. Currently, varieties of kinase inhibitors have been widely used in targeted therapy of a series of tumors and achieved clinical results. Radiomics is the bridge between medical imaging and personalized medicine. Quantitative analysis of imaging features provides not only the tumor phenotype but the underlying genotype information, which extends the analysis of imaging from qualitative to quantitative analyses and finds the clinical significance that cannot be found with the naked eye. The alterations in the levels of DNA, RNA, protein, metabolite, and medical imaging constructed the myriad of dysfunctionally mutually associated molecular networks making cancer be a complex systems biology disease [19,20,21]. Any individual study in a level is insufficient to clarify the intricate pathogenesis of a cancer. The integration of multi-omics data plays a pivotal role in elucidation of the molecular mechanism of tumorigenesis and discovery of new biomarkers and drug targets [19, 22]. Thus, a radical shift in cancer treatment is occurring in terms of predictive, preventive, and personalized medicine (PPPM) [23,24,25]. This review article describes basic principle, challenges, advances and clinical applications of different “omics” technologies, and highlights the significance of integrating multi-omics data in cancer research and in evaluating clinically relevant outcomes.
Methodology and application of genomics in cancer research and clinically relevant outcomes
Methodology
Since study found that the abnormal chromosome distribution during cancer cells division suggest a role in malignancy in 1914 [26], ones began to explore the connection between abnormal genetic substance and tumorigenesis. The in-depth studies of chromosome discovered Philadelphia chromosome that was resulted from the translocation between chromosome 9 and 22 in chronic myelogenous leukemia (CML) cells [27]. Since a seminal discovery of a single point mutation of HRAS (a guanosine was substituted to thymidine) that was responsible for the activation of oncogene in T24 human bladder carcinoma cells in 1982 [28], more oncogenes such as EGFR [9], RAS [29], PI3K [30], and ERK [31] have been recognized. Those findings promote scientists to increasingly understand cancers that are derived from accumulation of genomic alternations, including base substitutions, small insertions and deletions, chromosomal rearrangements and copy number alterations and microbial infections [32]. Less than 3 years after the completion of Human Genome Projects, the National Institutes of Health has officially launched the pilot stage of an effort to create a comprehensive catalogue of the genomic changes related to cancer in 2006, namely the Cancer Genome Atlas (TCGA) [33]. Moreover, the international Cancer Genome Consortium (ICGC) and the Cancer Genome Project of the United Kingdom share the same goals that identify all genomic alternations significantly associated with cancer.
The development of cancer genomics is inseparable from the progress of DNA sequencing technology. From the first-generation sequencing to the next-generation sequencing, DNA sequencing technology has developed by leaps and bounds. Here, the development of technologies in DNA sequencing is reviewed.
First, Sanger invented “the dideoxy method” in 1977 [34], which improved the method of the previous “plus and minus” [35, 36] for DNA sequencing. Sanger sequencing based on the selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication had been predominant method in this filed for almost 30 years [34, 37]. With long read lengths (up to ~ 1000 bp) and high per-base “raw” accuracies as high as 99.999% [38], Sanger sequencing achieved a number of monumental accomplishments, including completing of the Human Genome Project [37]. However, it has the obvious disadvantages of high cost and low throughput [3, 37]. The demand for entirely new technologies that deliver fast, inexpensive, and accurate genome information catalyzed the development of next-generation sequencing (NGS) technologies.
The second-and third-generation technologies are referred to as NGS [37]. By now, several commercially available platforms such as Roche/454, Illumina/Solixa, Life/APG, and Helicos BioSciences are all characterized by cyclic array sequencing summarized as the sequencing of a dense array of DNA features by iterative cycles of enzymatic manipulation and imaging-based data collection [38]. Parameters of partial platforms were summarized (Table 1). The advantages of second-generation sequencing relative to Sanger sequencing include the higher speed and throughput, cyclic array sequencing to provide with > 106 reads/per-array and lower cost, the relatively easier gene library construction, higher degree of parallelism, and more efficient use of reagents [38, 39]. The disadvantage that limited the application of these platforms are shorter read lengths with an average read length range from 32 to 330 bp [37]), which creates challenges for genome alignment and assemble [3, 37, 38, 40, 41]. In the aspect of raw accuracy, the NGS platforms are at least tenfold less accurate than Sanger sequencing [38]. In addition, the overall cost is still high, 1–60 dollar/megabase [38], although the cost per base is lower by several orders of magnitude compared to Sanger sequencing [39].
The third generation of sequencing technology such as PacBio RS and Oxford Nanopore sequencing is developed to solve the shortcomings of the second-generation [42], with fundamental feature of the single molecule sequencing but not requirement of any PCR process, which effectively avoids the PCR bias caused by the system error, improve the read length, and maintain the advantages of high-throughput and low cost of the second-generation technology.
Application
All cancers arise as a result of changes that have occurred in the DNA sequence of the genomes of cancer cells [43]. Thus, discovery of new somatic mutations, especially the “driver gene” mutations, has been at the heart of cancer research for more than a century. With the application of the NGS, identification of all genomic abnormalities in cancers has been turned from fantasy into reality. TCGA research network has showed the comprehensive genomic characterization of squamous cell lung cancers [44], gastric adenocarcinoma [45], human colon and rectal cancer [46], human glioblastoma [47], and ovarian carcinoma [48]. The study of lung squamous cell carcinoma (LSCC) found a mean of 360 exonic mutations, 165 genomic rearrangements, and 323 segments of copy number alteration per tumor, and loss-of-function mutations that are not reported previously. Besides, a potential therapeutic target was identified to offer new avenues of investigating the treatment of LSCCs [44]. Up to date, many types of cancers have been sequenced with whole genome sequencing (WGS) or targeted genome sequencing (Table 2) [7, 49,50,51,52,53,54,55,56,57,58].
The application of high-speed and high-throughout NGS technologies improves significantly the analysis of cancer genome, and reveals the full repertoire of mutated cancer genes, which not only can be used to guide the discovery of new targeted drugs, but also have an overwhelming impact on understanding of cancer biology and accelerate strategies in PPPM in cancer. For example, gene fusions resulting from chromosome translocations have an important role in the initial steps of tumorigenesis with evidence of discovery of gene fusions in all malignancies [59]. Functionally recurrent gene fusions provide more precisely clinical-related subclassifications of traditionaly morphological classification of tumors and accelerate the development of specific targeted therapies. Previously, because of lacking systematic approaches, this type of molecular abnormality has been regarded as a fundamental mechanism in haematological and soft-tissue malignancies. Recent years, with the application of NGS, novel recurrent chromosomal rearrangements have been discovered in many kinds of solid tumors, such as TMPRSS2-ETS fusion oncogenes in prostate cancer (Pca) [60], EML4-ALK fusion oncogenes in non-small cell lung cancer (NSCLC) [61], ETV6-NTRK3 fusion oncogenes in secretory breast cancer [62], BRAF and RAF1 fusion oncogenes in melanoma [63], BRAF gene fusions in pilocytic astrocytomas, pancreatic acinar and papillary thyroid cancers [64]. By July 2017, the Tumor Fusion Gene Data Portal (http://www.tumorfusions.org/) has presented 33 tumor types and a total of 20731 fusion genes information. The common fusion genes are kinase and transcription factors, which play an important role in tumorigenesis and metastasis and shed light on the PPPM practice in cancer [65]. Some clinical studies have evaluated the diagnostic and prognostic values of TMPRSS2-ERG gene fusion for Pca, which demonstrated that TMPRSS2-ERG had prognostic value and its combination with prostate cancer antigen 3 (PAC3) can increase the precision of PSA-based diagnosis [66, 67]. More importantly, the character that TMPRSS2-ERG gene fusion could be measured in the urine makes it an ideal biomarker supplementing the PSA test [67, 68]. ETV6-NTRK3 fusion oncogene was discovered in 90% secretory breast carcinoma (SBC), a rare subtype of infiltrating ductal carcinoma, but not in other ductal carcinomas [62]. In addition, ETV6-NTRK3 fusion oncogene was also reported in a rare salivary gland tumor similar to SBC leading to a newly described type of salivary carcinoma-secretory carcinoma (SC) [69]. Studies demonstrated that ETV6-NTRK3, a chimeric protein tyrosine kinase, depended on insulin-like growth factor 1 receptor signaling and induced insulin-receptor substrate-1 (IRS-1) constitutively tyrosine phosphorylated and consequently activated Ras-Erk1/2 and PI3K-AKT signaling pathways during transformations [70, 71]. Functional studies suggest these cells and cancers may sensitive to kinase inhibitors. A pan-NTRK as well as ALK and ROS1 tyrosine kinase inhibitor, entrectinib, has been found useful in treating a single patient with SC, which demonstrated the potential role of kinase inhibitor in treating of ETV6-NTRK3 fusion gene-associated cancers [72]. EGFR mutants were the most common genomic alteration underlying NSCLC, and patients with EGFR mutants were routinely treated with EGFR kinase inhibitor. Recent years, new recurrent fusion oncogenes EML4-ALK and FGFR3-TACC3 have been identified in NSCLC [61, 73]. These forms of molecular abnormalities have distinct mechanisms of tumorigenesis from EGFR mutants. The former is sensitive to ALK tyrosine kinase inhibitors such as crizotinib (approved by FDA in 2011) and the latter to fibroblast growth factor receptor (FGFR) kinase inhibitors such as BGJ398 (under clinical trials) [73, 74]. These findings complement the genotyping diagnosis of NSCLC and will benefit specific types of patients, ultimately enabling personalized medical treatment.
Methodology and application of transcriptomics in cancer research and clinically relevant outcomes
Methodology
The genetic central rule shows that genetic information is transferred from DNA to protein through RNA (mRNA) under precise regulation. The mRNA is regarded as a “bridge” in the process of biological information transfer from DNA to protein. Transcriptome is whole intracellular transcripts and their quantity in a given time and environmental condition. Transcriptome is an essential objective to address the functions of genome, uncover the molecular constituents of cells, and reflect the occurrence and development of a disease. The key aims of transcriptomics are to catalogue all species of transcripts, denote the transcriptional structure of gene, and quantify the expression level of each transcript during development and under different conditions [75]. Unlike genome that is a relatively static entity, transcriptome is dynamic, and modulated by external and internal factors. Therefore, transcriptome serves as a dynamic link between an organism’s genome and its phenotype characteristics [76].
Up to now, various methods have been developed to study transcriptome, including hybridization-or sequence-based approaches [75]. The former is based on hybridization between nucleic acids, which typically involves incubation of fluorescently labeled-cDNA derived from reverse transcription of different mRNAs with microarrays that are consisted of genes of interest, followed by digitalization with the specialized scanner and image analysis. Information is achieved such as gene name, clone identifier, and intensity values [77]. Recently, tiling microarrays derived from the standard gene expression microarray are composed of oligonucleotide probes that span the entire genome of an organism to provide a more unbiased view of the transcriptional activities within a genome [78]. However, several shortages of these methods include the reliance on existing knowledge of genome sequence, high background levels owing to cross-hybridization, and a limited dynamic range of detection due to both background and saturation of signals. Sequence-based approaches determine cDNA sequence but not rely on the probes. The sequences of cDNA or EST libraries were initially detected by Sanger sequencing approach; however, it is relatively expensive, low throughput, and generally no quantitative information. Afterwards, tag-based methods were developed to overcome those limitations, including serial analysis of gene expression (SAGE), cap analysis of gene expression (CAGE), and massively parallel signature sequencing (MPSS), which can provide high throughput, and precise gene expression levels, but are still based on Sanger sequencing technology that results in an analysis of only a portion of the transcripts and indistinguishing isoforms. The emergence and development of NGS provides a new approach, RNA-seq, for this high-throughput DNA sequencing technique in mapping and quantifying transcriptome (Fig. 2). The advantages of RNA-Seq include (1) high throughput, namely RNA-seq can achieve several to hundred billion of base sequences, which can cover the entire genome or transcriptome; (2) high sensitivity, namely RNA-seq can detect only a few copies of rare transcripts in a cell; (3) high resolution, namely RNA-Seq can achieve single-base resolution with good accuracy and avoid the level of high background; and (4) no reconstructions, namely RNA-seq can be used for the analysis of whole transcriptome of any species, including detection of unknown genes or transcripts, and accurate identification of the cleavage site, and a variable SNP or UTR region.
Application
Alternative splicing of precursor messenger RNA from a single gene was first discovered about 30 years ago, which produces multiple different functional messenger RNAs, and the corresponding proteins derived from the a single gene [79]. Splicing abnormalities are a common characteristics of cancer [80], occurring in every category of cancer hallmarks [81]. Abnormal splicing could result in aberrant protein variants to involve different functions such as transcription factors, cell signal transducers, and components of the extracellular matrix [82]. The nature of the altered gene products is usually consistent with an active role in cancer. RNA-seq can directly and readily detect RNA splicing events relative to standard gene expression microarray, so it is a power tool in discovering cancer-related alternative splicing, which might be a diagnostic or prognostic marker and potential personalized therapy target.
In the research of NSCLC, a comprehensive study of prognosis-related alternative mRNA splicing using RNA-seq data identified a large number of alternative splicing events that are associated with the prognosis of NSCLC. Furthermore, prognostic predictors based on alternative splicing events were established for risk stratification with excellent performance [83]. RNA-seq also allows quantitative study of alternative splicing. Owing to alternative splicing, the insulin receptor has two isoforms: insulin receptor isoform A (IR-A) and insulin receptor isoform B (IR-B) [84]. Another study used bioinformatics methods to analyze RNA-seq data of both isoforms found that downregulated IR-B level and increased IR-A/IR-B mRNA ratio correlated with lower epithelial-mesenchymal transition and longer survival time. In addition, this phenomenon has been found in other 18 types of cancers, which suggests this ratio could be used as a marker of prognosis and treatment response assessment [85]. In breast cancer, several EMT-associated alternative splicing events have been identified and most of these alternative splicings are regulated by one or more members of splicing factor classes such as PBFOX and ESRP, which may provide new diagnostic and prognostic markers and personalized treatment targets of a breast cancer [86].
Compared to the analysis of DNA sequencing-based structural variations, transcriptomics can provide with an analysis of DNA functional characteristics in the RNA level to link the gene structural feature to its functions and easier discover the causal of physiological or pathological conditions [87, 88]. RNA-seq has been proved to be a useful tool for the discovery of new gene fusions in cancer transcriptome. For example, one rather common and tumor-specific novel fusion gene SYT8/TNNI2 was discovered in analysis of three bladder carcinomas with high-throughput RNA-seq, which has potential clinical relevance [89]. Also, oncogenic gene fusions were revealed systematically in primary colon cancer with IIumina RNA-seq, with a result of a relevant gene fusion occurring 2.5% of all specimens; of them, USP9X-ERAS formed by chromothripsis was considered as highly oncogenic, with the ability to activate AKT signaling [90]. The analysis of ovarian cancer RNA-seq data with a novel computational method for fusion discovery—deFuse provides the first gene fusion discovery of ovarian cancer, which may contribute to the study of tumor initiation, development and treatment [91].
Micro RNAs are short (~ 22 nucleotides in length) non-coding RNAs (ncRNAs) that regulate gene expressions by binding to specific mRNA targets and promoting their degradation and/or translational inhibition [92]. Recent studies suggest that miRNAs play roles in cancer [93,94,95,96,97]. RNA-seq is a powerful tool to uncover unannotated ncRNA species. The abundant expression of miRNA-1323 and its distinct association in tumors arising from a cirrhotic background were discovered in hepatocellular carcinomas (HCCs) [98], and overexpression of miRNA-1323 in cirrhotic-HCCs was correlated with poorer disease-free and overall survivals of patients. In the study of myelodysplatic syndromes, the analysis of RNA-seq data demonstrated that the expression of miRNA was associated with the progression of the disease [99]. The miRNA-mRNA regulatory network was studied in peripheral blood mononuclear cells of small cell osteosarcoma (SCO) with RNA-seq [100], which identified 37 dysregulated miRNA (27 upregulated and 10 downregulated) and 1636 dysregulated mRNAs (555 upregulated and 1081 downregulated), two important signaling pathways including mTOR signaling and cell cycle signaling, and dysregulation of three miRNAs (has-miR-26b-5p, has-miR-221-3p, and has-miR-125b-2-3p) that might be involved in SCO tumorigenesis.
In addition to miRNAs, a large proportion in a transcriptome is long ncRNAs (lncRNAs) with longer than 200 nucleotides, which are often polyadenylated and are devoid of evident open reading frames these [101]. Studies demonstrate that lncRNAs are able to regulate gene expressions at the levels of chromatin modification, transcription, and post-transcriptional processing [101, 102], especially in some human cancers with tissue-specific expressions [103], demonstrating their potential roles in both oncogenic and tumor-suppressive pathways [104, 105]. Currently, the study of lncRNAs is still in its initial stage with studies of only a small part of lncRNAs such as HOTAIR [102, 106], and MALAT1 [107, 108]. However, IncRNAs demonstrate its big potential in PPPM practice, and RNA-Seq is maximizing the coverage of cancer-related lncRNAs in a transcriptome. For example, among 121 unannotated prostate cancer-associated ncRNA transcripts, PCAT-1 was discovered as a prostate specific regulator of cell proliferation and a transcriptional repressor in a subset of prostate patients [109]. RNA-seq systematically identified quintuple-negative lung adenocarcinoma-related IncRNAs [110], including 90 upregulated and 153 downregulated lncRNA transcripts. The functions of 14 predicted lncRNAs such as vasculature development and cell cycle are closely related to the process of cancer development. Another study [111] identified a signature of five lncRNAs (CYP4F26P, RP11-108M12.3, RP11-38M8.1, RP11-54H7.4 and ZNF503-AS1), which might act as an independent prognostic indicator for LUSC with RNA-seq data from TCGA. Similarly, a signature of eight lncRNAs was identified to stratify and predict survival in esophageal cancer [112].
Methodology and application of proteomics in cancer research and clinically relevant outcomes
Methodology
Proteins are the effectors of DNAs in a biological system, and the expression levels of all proteins in a proteome would inarguably provide the most relevant phenotype characteristics of that biological system [113]. The goal of proteomics is to characterize information flow with protein pathways and networks to eventually understand the function relevance of proteins in a cell or organism [4]. The proteome has many unique features that distinguish from other omics approaches, and is much more complex than genome and transcriptome. The number of human proteins and their variants or protein species is estimated up to over billions [19]. Also, one gene corresponds to multiple proteins, namely one gene-multiple proteins model but not one gene-one protein model [114, 115]. In addition, variations in a proteome are more measureable than variations in genome and transcriptome [116]. It seems that genome contains all information; however, except for the sequence and copy number of DNAs and RNAs, other information in a genome is difficultly measured with current technologies. Proteome as an important component of a phenome is the final performer of genome functions; much information in a proteome is measurable such as amino acid sequence, splicing, copy number, post-translaitonal modifications (PTMs), variants, spatial conformation, and spatial re-distribution. In the last decade, numerous proteomics studies have focused on protein profiling and protein expression alternations that associate different given conditions.
Proteomics method commonly includes protein preparation, separation, and identification (Fig. 3). Protein separation is to reduce the complexity of the proteome sample, mainly includes gel- and liquid chromatography (LC)-based approaches. The gel methods include one-dimensional gel electrophoresis (1DGE), two-dimensional gel electrophoresis (2DGE) [117], and two-dimensional difference in gel electrophoresis (2D-DIGE) [118]. The specific antibody must be used in combination with those gel methods if variants of a given protein [118], or a kind of PTM [119,120,121] need to be detected. The LC methods as proteomic separation technique are extensively used in the field of current proteomics, mainly include 2DLC and multi-dimensional LC (MDLC), and a stable isotope (e.g. iTRAQ and TMT) labeling coupled with 2DLC can quantify the component of a proteome. Moreover, some LC methods in combination with MS are developed to identify protein variants, and protein species [122,123,124,125,126]. MS is the key protein identification technique, which can determine amino acid sequence of a protein [115], and PTM-sites [120]. Different types of mass spectrometers are commercially available, including matrix-assisted laser desorption ionization-time of flight-time of flight (MALDI-TOF-TOF)[127], Fourier transform ion cyclotron resonance (FTICR) [128, 129], triple TOF 5600 or 6600 systems [130], and LTQ orbitrap system [131, 132] with different ion fragmentation models such as collision induced dissociation (CID) [133], electron transfer dissociation (ETD) [134], and electro capture dissociation (ECD) [128, 135], which provides the optimal strategies to identify protein expressions, PTMs, protein variants and protein species. However, one must realize that each mass spectrometer has its own sensitivity and resolute capability, an enrichment strategy is needed prior to MS in analysis of low abundance protein, PTMs, or protein variants [126, 136].
MS-based proteomics includes top-down and bottom-up approaches. Top-down proteomics is able to identify and quantify unique proteoforms through feeding intact full proteins directly into MS, which is capable of providing distinct characteristics of each kind of proteoform with more precise and more abundant biological information [137]. Bottom-up proteomics digests firstly protein components with enzyme, followed by LC fractions and MS-identification, which is able to identify and quantify proteins expressed differentially, and PTMs [138]. Recently, middle-down method that combined top-down and bottom-up strategies receives attentions in that this method not only can avoid redundant peptides sequences but also can analyze large protein fragments [139].
Quantitative proteomics plays very important roles in understanding the biological significance, mainly including 2DGE-based quantitative methods [140, 141], stable isotope-labeled quantitative methods such as isobaric tags for relative and absolute quantification (iTRAQ) [142, 143], and label-free quantitative methods [144, 145] such as selected/multiple reaction monitoring (SRM/MRM) [146, 147], and sequential window acquisition of all theoretical mass spectra (SWATH) [148, 149]. Furthermore, structural proteomics benefits in-depth understanding of the biological functions of a protein in a biological system [150, 151].
Application
Discovery of new tumor biomarkers is the hot point in the field of cancer research with high-throughput MS-based proteomics. For example, glycosylated proteins represented 50% of the secreted proteome and abnormal glycosylation of proteins has been implicated to play a critical role in cancerous progression [152]. Since more than half of the proven cancer biomarkers are glycosylated proteins, MS-based glycoproteomics can analyze qualitatively and quantitatively thousands of glycosylated proteins with detailed information, which shows a great potential in discovery of novel cancer biomarkers. Thus, glycoproteomics has extensively used in cancer research. Several examples are taken here.
Quantitative proteomics analysis of fucosylated glycoproteins in small cell lung cancer (SCLC) patients [153] found a significant decrease of PON1 protein expressions in the sera of SCLC patients, but a significant increase of PON1 fucosylation. The altered fucosylated glycan patterns and levels of PON1 were used as potential diagnostic and prognostic biomarkers for SCLC. Another MS-based glycoproteomics identified the significantly increased fucosylated haptoglobin (HP) with three α-2, 6-linked sialic acids, in serum of each subtype of lung cancers (19 lung adenocarcinoma, 8 LSCC, 11 SCLC and 7 unknown types) relative to controls [154]. This specific glycan of Hp from the serum can serve as a potential diagnostic glycobiomarker for lung cancer.
Glycoprotein biomarkers were also studied in HCCs. Compared to liver cirrhosis patients, an integrated approach analyzing glycoproteins and their glycosylations in HCC sera found the significantly increased levels of 5 fucosylated glycoproteins, which can be regarded as early diagnostic biomarker candidates with excellent performance [155]. Also, AFP-L3, which is an isoform of AFP, and binds strongly to lens culinaris agglutinin (LCA) by an additional α1-6 fucose residue at the reducing terminus of N-acetylgucosamine, has been determined as an early and highly specific biomarker for HCC with sensitivity 56% and specificity 95% [156].
Quantitative glycoproteomics has been used to study Pca with a high incidence and low mortality [157,158,159]. Prostate-specific antigen (PSA) was an FDA approved serum biomarker for Pca diagnosis and prognosis with low specificity, and cannot distinguish aggressive Pca from non-aggressive Pca, which might result in overtreatment of non-aggressive Pca patients. To obtain the urgently needed novel biomarker for Pca patients, SWATH-based glycoproteomics discovered and validated two glycoproteins (N-acylethanolamine acid amidase, and protein tyrosine kinase 7) in Pca tissues as Pca aggressive biomarkers [160], which provides a basis for the precise treatment of Pca patients, and reduces side effects of Pca overtreatment.
In addition to glycosylation of proteins, other types of PTMs in proteins also constitute a large number of diagnostic and prognostic biomarker candidates. For example, phosphoprotein secretomics studies provided a set of novel breast cancer subtype specific phosphopeptide candidates in plasma [161]. PGRMC1 is a membrane-related progesterone receptor and an important biomarker for breast cancer progression. Since phosphorylated PGRMC1 will active a series of intracellular signaling, it is a potential therapeutic target [162]. Based on tissue phosphoproteomics method in NSCLCs, PTRF/cavin-1 and MIF have been regarded as new potential biomarkers [163]. Protein tyrosine nitration is another important PTM, which changes the chemical properties of that tyrosine residue and protein functions [151, 164]. 2DGE-based nitroproteomics [119] identified 18 nitroproteins and 20 nitrotyrosine sites in human high-grade astrocytomas, which are associated with a series of biological processes such as drug assistance and signal transduction, provide new insights into pathogenesis of astrocytomas, and benefit the discovery of new biomarkers for its early diagnosis and effective therapeutic targets [165].
Besides biomarkers, proteomics approach is also a guiding tool for the discovery of more potential therapeutic targets, for example, BIRC6 in colon cancer stem cells [166], bone marrow stromal antigen 2 and cyclophilin A in endometrial cancers [167, 168], phosphoglycerate mutase 1 in HCCs [169], anaplastic lymphoma kinase in ovarian cancer [170], and hypusination of eukaryotic initiation factor 5A in BCR-ABL-positive leukemias [171].
Above examples are only windows for the use of proteomics in cancer research. Here, one must realize that the initiation and development of each types of tumor are related to a distinct series of molecular pathogenic defects. Personalized treatment of cancer requires dynamic monitoring the whole abnormal molecular events and interaction among them. MS-based proteomics and pathway network analysis tools have become an essential approach in accelerating personalized treatment. For example, pathway network analysis based on multiple sets of pituitary adenoma proteomics data (DEP data, nitroproteomics data, and protein mapping data) revealed mitochondrial dysfunction, oxidative stress, cell cycle dysregulation, and MAPK-signaling abnormality were significantly associated with pituitary adenoma pathogenesis [172], wich provides new clues to in-depth investigation of pituitary adenoma and discovery of effective biomarkers. Another protein-protein interaction (PPI) analysis of HCCs depicted the molecular portrait and revealed the relationship among metabolism, cytoskeleton biological processes, and HCC metastasis [173].
Methodology and application of metabolomics in cancer research and clinically relevant outcomes
Methodology
Metabolism is one of the key components of life. Studies have shown that the physiological state of cells and tissues is determined by both the cell’s regulatory systems and its state of intermediary metabolism [174]. Metabolites are small molecules (< 1 KDa) derived from metabolism, and provide functional information that cannot be directly obtained from genome and proteome of the cellular and tissue states [175, 176]. These metabolic profiles are associated with totally biochemical processes as beginning, intermediate, or end products and provide information on complex interactions between genes and environment of a given condition [177, 178]. Also, metabolites can feed back on other physiological and pathological processes [179,180,181,182]. Metabolome contains all endogenous metabolites and is divided into primary metabolome (governed by the host genome) and co-metabolome (dependent on the microbiome) [175]. Metabolome-wide association is able to uncover the etiology decided by the intricate interaction of genes, environment and lifestyles in the general population [183]. Metabolomics is the methodology and theory to comprehensively and dynamically study metabolome [184], including identification biochemical and molecular characteristics of metabolome, characterization of interactions among different metabolites or between metabolites and genetic/environmental factors, and evaluation of biochemical mechanisms related to a given condition such as different pathophysiological processes [185]. In general, metabolomics can be divided into targeted metabolomics and untargeted metabolomics. Targeted metabolomics refers to a method where a specified list of metabolites is measured, typically focusing on one or more related pathways of interest. Targeted metabolomics is commonly driven by a specific biochemical question or hypothesis that motivates the investigation of a particular pathway [176]. Untargeted metabolomics is a globally and simultaneously measurement of as many metabolites as possible from biological samples without bias [176].
NMR spectroscopy (mostly proton NMR, 1H-NMR) and chromatography coupled to MS (LC-MS and GC-MS) are two leading spectroscopic techniques used in metabolomics [186]. Numerous favorable characteristics make NMR a beneficial tool in metabolomics research. NMR-based methods have high reproducibility in the laboratory and between laboratories [187,188,189]. NMR enables the identity of structures for unknown metabolites [190,191,192] and possesses the ability to non-constructively analyze samples that do not need to separate and elaborately prepare samples, which could be analyzed subsequently with other platforms [193,194,195,196]. Moreover, with isotope labeling, NMR provides a window to observe the dynamic changes of metabolite formation and metabolic pathways, which could be used to follow the perturbation of metabolites before and after intervention treatment [197, 198]. Since the 1970s, chromatographic methods have been used to separate complex mixture of metabolites and improve analysis and identification [199]. GC and GC-MS methods have been used to quantify metabolic profiling, but GC-MS is largely limited to volatile compounds [199]. LC-MS has significantly improved the capability of MS-based metabolomics because it is more sensitive than 1HNMR and can identify and quantify a few hundred metabolites within a single extract [199, 200]. However, each method has its own advantages and disadvantages. NMR is less sensitive than MS by up to 100-fold, and the instrument is expensive. LC-MS is highly sensitive, but it is necessary to separate and prepare samples, which might potentially modify metabolite structure to increase the difficulty in analysis. None of them alone can effectively identify and quantify, with sufficient sensitivity and precision, the diverse range of metabolites and their dynamic changes in cells. An integrated method of these methods is necessary to increase the accuracy and efficiency of identification of those metabolites and benefit the development of metabolomics [201]. The characteristics of NMR, GC-MS, and LC-MS, and the examples of applications in cancer research were presented (Table 3).
Application
Cancer is involved in a range of metabolic process changes. Metabolites are the products of the interactions between genes and environment. The metabolites are closer to the phenotype of the organism than genes and proteins. Early diagnosis is critical to improve the survival of cancer patients. Metabolomics is considered as a relatively rapid, accurate and noninvasive method, it is becoming an increasingly popular tool in discovery of diagnostic biomarkers of cancers [209, 210]. Many enthusiastic metabolomic markers have been reported for diagnosis and prognosis in lung cancer [205, 208, 211], breast cancer [204, 212], pancreatic cancer [206], Pca [213,214,215], bladder cancer [216,217,218], and epithelial ovarian cancer [219, 220].
For example, metabolomics has been used to discover noninvasive diagnostic biomarkers for lung cancer with high incidence and mortality. The unbiased LC-MS analysis of the metabolic profiling of urines from 469 lung cancer patients and 536 controls [208] revealed creatine riboside and N-acetylneuraminic acid (NANA) were the powerful urinary clinical metabolomic biomarkers for putative diagnosis and prognosis, which was further confirmed in an independent population with 80 patients and 78 controls. Also, sweat metabolomics was used to discover noninvasive biomarkers for diagnosis and prognosis of cancers. LC-MS analysis of metabolome of lung cancers relative to normal smokers identified trisaccharide phosphate as an individual metabolite biomarker to discriminate lung cancer from controls with the specificity of 80% and sensitivity of 72.7% [211], and a panel of five metabolites (trihexose, tetrahexose, suberic acid, monoglyceride MG (22:2), and nonanedioic acid) significantly improved the specificity (80%) and sensitivity (79%). Moreover, the sputum metabolomics analysis [205] between 34 lung cancer patients and 33 healthy controls found that ganglioside GM1 might be a reliable candidate for biomarker and showed that sputum metabolomics method could help ones to screen the high-risk population of lung cancer.
Metabolomics has also been used in breast cancer research. UPLC-MS/MS analysis of saliva metabolite profiling of breast cancer patients identified the ratios of polyamines, eight polyamines, as noninvasive diagnostic biomarker to effectively discriminate breast cancer patients from healthy controls [212]. GC-MS analysis [204] of serum metabolomes of 152 pre-operative breast cancer patients and 155 healthy controls identified seven metabolites (tetradecane, alpha-D-glucopyranoside, methylstearate, dodecane, 1-4-benzene, D-galactose, and octadecanoic acid) that were significantly associated with breast cancers, found metabolic content differs between cancer and benign tissues, and also identified differentiated metabolites for grading, staging and determination of neoadjuvant status.
MS-based metabolomics [206] revealed four metabolites (oleanoic acid, taurochenodeoxycholate, palmitic acid, and d-sphingosine) as highly discriminative potential prognostic markers for pancreatic cancer, a poor prognostic cancer with 5-year survival rate < 5%, demonstrated that palmitic acid has a better discriminating ability compared to the CA19-9 that is only biomarker routinely used for the clinical management of pancreatic cancer, and recommended simultaneous assessment of palmitic acid and CA19-9 to reduce false positives and improve prognosis of patients. It suggests metabolomics plays an important role in prognosis research of pancreatic cancer.
The increase of efficiency and decrease of the side effects in cancer therapy have always been the focus of cancer research, which is actually consistent with the goal of precise medicine that is to use advanced multiomics testing to customize a personalized medical treatment according to their specific biomarker profiling. Cancer genomic profiling is now routinely used to guide the cancer precision medicine, and made some achievements. However, the heterogeneities of cancer tissues and cancer genomes make it impossible alone to guide precise treatment of cancer. Genomic profiling is a powerful tool to provide the information what will happen in tumor, whereas metabolomics can provide the information what has happened and is happening in cancer. Metabolomics has the ability to measure the sum of all these genotypic, environmental and physiological effects, thus it is a very promising method for the use of metabolomics to predict and assess responses to anticancer treatments in cancer research, and it is possible for the use of metabolic profiles to predict the response of individual patients to a class of treatments.
For example, the untargeted serum metabolomics of lung adenocarcinoma patients before chemotherapy identified and constructed a metabolite pattern model to predict the response of pemetrexed and platinum treatment demonstrating the metabolomics-based method is an effective approach to identify appropriate patients who are more likely to a special treatment [221]. Metabolomics analysis of human xenograft model of gastric cancer established a prediction model containing 1-acyl-lysophosphatidycholines, polyunsaturated fatty acids and their derivatives, which can predict the chemosensitivity of cisplatin plus 5-fluorouracil with an accuracy of 90.4% [178]. Similar metabolomics-based predictive studies were also carried out in other types of cancers [209, 219, 220]. Those examples clearly demonstrated that metabolomics is an effective method to stratify patients, establish reliable predictive models to predict the response of cancer patients before the treatment, and improve the efficacy and survival time of patients. Moreover, the immediately measurable metabolic perturbations are occurring in a large number of tissues after exposure to a particular antitumor agent, these metabolic changes represent a biomarker of efficacy or toxicity, which is easily detected by metabolomics methods. A 1H MRS-based metabolomics analysis of Degarelix that decreases serum androgen levels in human advanced Pca found that the degree of concentration decline of two metabolites (lactate and t-choline) was able to monitor noninvasively the response of castration [202]. The use of hyperpolarized MRI-based metabolomics to study of targeting PI3K/mTOR pathway in sarcomas found lactate was a biomarker to assess the treatment response to rapamycin [222]. Metabolomics also plays important roles in monitoring radiotherapy toxicity. The 1H NMR-based serum metabolomics analysis found the increased N-acetyl-glycoprotein and acetate was the biomarkers to reflect the acute radiation sequelae (ARS) in head and neck squamous cell carcinoma patients [203].
Those evidences clearly demonstrate that metabolomics method is more accurate and faster in assessment of treatment response compared to the traditional method such as imaging examination in evaluation of anticancer effects.
Currently the understanding of cancer is gradually shifted from a genetic disease to a metabolic disorder [223, 224] because metabolites not only reflect the metabolic state of cancer but also feedback the information on the occurrence, development, and consequence of cancer. With the extensive application of metabolomics technology in cancer research, a new term “oncometabolites” are proposed and defined as endogenous metabolites and their accumulation that initiates or sustains growth and metastasis of cancer [225]. A series of oncometabolites have been identified, including 2-hydroxyglutarate and glucose in gliomas and acute myeloid leukemia [226,227,228], fumarate in papillary kidney cancer [229], succinate in pheochromocytoma [230], sarcosine and choline in Pca [231, 232], glutamine in pancreatic [233, 234], asparagine in ovarian cancer [235], and lactate in breast cancer [236, 237]. Those oncometabolites are leading to identity of novel drug targets and therapeutics.
For example, isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) are critical metabolic enzymes that catalyze isocitrate to α-ketoglutarate. Mutated IDH1/2 was found a neomorphic enzymatic activity to catalyze α-ketoglutarate to (R)2-hydroglutarate [(R)2-HG] in gliomas [238, 239]. The accumulation of 2-HG inhibits 2-oxoglutarate-dependent oxygenases [240], impairs histone demethylation [241], blocks cell differentiation [242], and promotes tumorigenesis [243]. Tumor with IDH mutation constructs a distinct clinical subset in both leukemia and gliomas. IDH mutations were also identified in multiple cancers, including chondrosarcoma [244], sarcoma [245], and cholangiocarcinoma [246]. IDH mutants become promising candidates of therapeutic targets. A selective R 132H-IDH1 inhibitor (AGI-5198) demonstrated that mIDH1 inhibitor was able to block the production of R-2HG, and induce demethylation of histone and the expression of gliogenic differentiation associated genes, but it did not influence the functions of IDH1 wild-type in a glioma [247]. This inhibitor AGI-5198 also demonstrated the similar effects in human chondrosarcoma cells [248]. The IDH2 inhibitor AGI-6780 also induced differentiation of TF-1 erythroleukemia and primary human acute myelogenous leukemia cells [249]. More and more IDH inhibitors are being developed such as AG-120 [250] and AG-221 [251, 252] in cancers. Those studies clearly indicated that IDH mutations are targetable by small molecules, which provides a promising cancer therapeutic strategy, namely inducible differentiation therapy [253]. Inducible differentiation therapy is to reactivate endogenous differentiation programs, elicit tumor cell maturation, and transit cancer to normal tissue without cytotoxic effects, which can overcome drawbacks of traditional cytotoxic chemotherapy that is to inhibit and kill tumor cells with serious side effects [254]. The initial clinical application of IDH inhibitors, inducible differentiation agents, has demonstrated the strong potential in cancer therapy with minimal toxicity.
Therefore, those oncometabolites, IDH inhibitors and their clinical applications are the strong evidences in support of the importance of metabolomics technology in discovery of new anticancer drugs and therapeutics.
Methodology and application of radiomics in cancer research and clinically relevant outcomes
Methodology
Medical imaging technologies such as CT, PET/CT, and MRI play an irreplaceable role in the diagnosis and prognosis of tumors. In general, medical images are regarded as pictures. Physicians visually interpreted these “pictures” solely and draw qualitative and preliminary quantitative conclusions of tumors, including the location of tumor, internal heterogeneity, the overall and marginal morphology of the lesion, the relationship with surrounding tissues, rough measurements of diameter, the volume of tumor, CT and PET/CT values, MRI signal height and other values. This type of information is crucial for the diagnosis of tumors, but it does not accurately reflect the morphological and behavioral complexities of a tumor, with limited benefits in the judgment of treatment sensitivity and prognosis [255]. Whether one could further exploit the medical imaging to obtain the broader characteristics of tumor? In the past decade, medical imaging analysis and recognition technology has developed rapidly [256], which made it possible to extract and quantitatively analyze the entire information and spawned a new discipline-radiomics [257]. Radiomics, based on computer-aided diagnosis and detection systems, is defined as high-throughput extraction and conversion of quantitative features from medical imaging into mineable data and applied the analysis of these data within clinical decision support systems [256,257,258]. Since medical imaging is routinely used in clinical decision, radiomics, extending the imaging analysis from qualitative to quantitative and finding the clinical significance that cannot be found with the naked eye, may have a clinical impact on cancer research.
The general workflow of radiomics includes 4 steps (Fig. 4): (a) acquisition of high quality and standardized imaging, (b) identification of volumes of interest (VOI) and segmentation, (c) feature extraction and qualification, and (d) analysis and modeling. High quality and standardized imaging is the basic of radiomics. Unlike qualitative analysis, variations in acquisition and image reconstruction will jeopardize the ability to detect biological differences. So standardized imaging is important to eliminate unnecessary confounding variability. Segmentation determines which voxels within an image are analyzed, so it is the most critical and challenging component of radiomics. The ideal segmentation method should provide accurate and reproducible boundaries and should be time efficient, which means the entire process should be as automated as possible with minimum operator interaction. Myriad imaging features can be extracted and divided into tumor intensity histogram-based features, shape-based features, and texture-based features. Only those task-specific features have been selected and analyzed. Ideally, the final model based on selected features and methodology must be internally and externally validated.
Application
Radiomics, like the other omics, has equivalent potential role in PPPM of cancer. Several studies suggested the potential associations between certain radiomics features and tumor phenotypic patterns [259,260,261]. Analysis of radiomics-based features, comprehensive quantification information relating to the tumor phenotypes could be obtained [262, 263]. Moreover, potential noninvasive imaging biomarkers for prediction of treatment response and outcomes could also be provided. For example,a PET/CT imaging study in NSCLC showed that abnormal texture as measured by coarseness, contrast, and busyness is associated with nonresponse to chemoradiotherapy and with poorer prognosis [264]. Another study exploring a set of 635 CT-derived imaging features, including intensity, shape, texture, Laplacian of Gaussian, and wavelet filters, found that 35 and 12 features were related to distant metastasis and survival, respectively [265]. The utility of MRI texture features in glioblastoma demonstrated good performance (area under ROC curve > 0.7) in distinguishing different molecular subtypes and predicting 12-month overall survival status (area under ROC curve = 0.69) [266]. Similarly, based on a series of MRI imaging features of 81 patients, a prognostic model was established that has a potential role in guiding personalized treatment selection [267]. In Pca, Haralick texture analysis of prostate MRI has the ability to detect the tumor lesions and differentiating Pca with different Gleson scores [268]. Another study assessed T2-weighted MRI-derived textural features demonstrated that these features corrected significantly with Gleason score and could distinguish Gleason score 3+4 from 4+3 cancers with high sensitive to the pathological difference [269]. There are similar researches in esophageal cancer [270, 271], rectal cancer [272], breast cancer [273, 274] and head and neck cancer [275, 276]. In addition, radiomics could be used to predict radiotherapy-related side effect and guide personalized radiotherapy treatment. For example, the intensity and textural features based on CT of pre- and post-radiation therapy was analyzed in the study of the relationship between radiation dose and the development of radiation pneumonitis. As a result, 12 features showed a significant correlation with pneumonitis [270]. A similar study also found that texture features extracted from CT of nasopharyngeal cancer could be used in predicting parotid shrinkage at the end radiation therapy.
Furthermore, radiomics has distinct characteristics. In the era of precision medicine, genotype of tumor is an important basis for personalized treatment. Due to the high heterogeneity of tumor, the genomic profiling obtained from clinical biopsy is insufficient to reflect the real genomic state of a tumor. Simultaneously, not all cancer patients can undergo biopsy that may induce serious complications. In contrast, almost every cancer patient has radiologic images and radiomics could objectively and precisely provide detailed quantitative features of intra- and intertumoural heterogeneity in a non-invasive manner. Based on the hypothesis that genotypic variation is the source of a proportion of radiomic features variance, a new interdisciplinary radiogenomics mining of radiomics data to detect correlations with genomic patterns has been proposed. Radiogenomics facilitates an in-depth understanding of tumor biology and captures the intrinsic tumor heterogeneity and could provide diagnostic and prognostic imaging biomarkers to guide the precisely personalized treatment [277, 278]. For example, a study of 10 glioblastoma MRI features discovered that the ratio of enhancing to nonenhancing volume was correlated with EGFR overexpression. The enhancing phenotype was correlated with angiogenesis and tumor hypoxia-related genes [259]. Another glioblastoma study based on MRI-derived tumor imaging features demonstrated that TP53 mutant tumors had smaller enhancing and necrotic volumes (p = 0.012 and 0.017, respectively) and RB1 mutant tumors had smaller edema volumes (p = 0.015) [279]. A study of HCC found that microvascular invasion (MVI), an independent predictor of poor outcomes that cannot be adequately determined before operation, has very important clinical decision significance. In a study of contrast-enhanced computered tomography features of 157 HCC patients, venous invasiveness based on three features (internal arteries, hypodense halo and tumor-liver difference) was identified as a radiogenomic biomarker of MVI derived from a 91-gene HCC “venous invasion” gene expression signature. This biomarker has a good performance in detecting MVI with diagnostic accuracy of 89%, sensitivity of 76%, and specificity of 94%, respectively. Patients with a positive RVI score were associated with low overall survival than patients with negative RVI score in the overall cohort [280]. A study of cholangiocarcinoma in exploring of the relationship between imaging feature and hypoxia markers suggested that both qualitative and quantitative imaging features (based on texture analysis of CT) were correlated with a few hypoxia markers, such as VEGF, EGFR, and CD24 [281]. A study of breast cancer by combining radiogenomics with RNA-seq identified the enhancing rim fraction score, a quantitative dynamic contrast material-enhanced MR imaging IncRNA radiogenomic biomarker, which was associated with metastasis and expression of the known predictor of metastatic progression, HOTAIR [282]. Another potential advantage of radiomics is to identify breast cancer molecular subtypes that are crucial in personalized treatment and no low-cost genetic testing is readily available. For example, a multivariate analysis of relationship between 56 routine MRI-based imaging features (including morphologic, texture, and dynamic features) and molecular subtype demonstrated a strong association between the collective imaging features and both luminal A and Iuminal B molecular breast cancer subtypes. No association was found for either HER2 or basal molecular subtype and the imaging features [283]. Similarly, using the computer-extracted MRI image-based features of 91 biopsy-proven invasive breast cancers from TCGA/TCIA, a classifier model was established and evaluated with receiver operating characteristic analysis, which shown the ability to distinguish between molecular prognostic indicators. This study shows promise for high-throughput discrimination of breast cancer subtypes and may yield a quantitatively predictive signature of advancing precision medicine [284].
The integration of multi-omics data in cancer research and clinically relevant outcomes
Cancer is a complex disease and involves deregulation in different levels of DNA, RNA, protein, and metabolite; and those different levels of molecules are mutually associated [19, 22, 23, 116]. Any individual study in a different level is insufficient to clarify the intricate pathogenesis of a cancer. Integration of multiple omics data is essential to cancer research and fits the reality of a cancer [19], which will provide a holistic view of what really happened during normal cell malignant transformation and tumor progression, and have the potential in improvement of targeted therapy and the effectiveness of traditional therapies, in clarification of molecular mechanisms of cancer therapeutic resistance, and in discovery of novel biomarkers and targeted drugs.
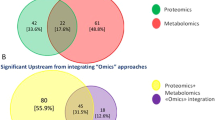
Integrated omics has been widely used in cancer research. For example, an integrated analysis of genomic and transcriptomic data and long-term clinical outcomes analyzing the changes of gene expression based on somatic gene copy number aberrations revealed some potentially important targeted therapeutic response-related events and proposed a new molecular classification of breast cancer patients [285]. Another integrative analysis of genomic and proteomic data demonstrated that PI3K pathway aberrations are particularly common in hormone receptor-positive breast cancer, which might be important in clinical selection of targeted therapies [286]. The integrated analysis of tissue transcriptomics and urine metabolomics identified four urinary biomarkers that are more credible compared to biomarkers derived from single omics [287]. The integrative analysis of transcriptomics, proteomics, and clinical outcome in HER2-positive breast cancers who acquired resistance to lapatinib revealed EGFR/HER2 signaling was still blocked, and the blocked intensity was weakened by the upregulation of glucose metabolism and endoplasmic reticulum stress pathways [288]. An integral analysis of transcriptomic and proteomic data in glioblatomas revealed a highly significant enrichment of gonadotropin-releasing hormone (GnRH) signaling pathway that was not deciphered with single omics datasets, which demonstrated the promise of multi-omics research and analyses to better understand complex cancers [289]. Moreover, an integrated quantitative proteomics and phosphoproteomics analysis was also used in sorafenib-treated failure HCCs and revealed that this targeted drug can indeed effectively inhibit its target kinase in Raf-Erk-Rsk pathway, but the downstream targets of Rsk-2 (eIF4B, filamin-A and so on) were not influenced, which suggests another alternative pathways might have been active and contribute to the treatment failure [290].
Outlook
The development of multiomics technologies benefits in-depth understanding of tumor biology. However, it is still very challenging in translating those multiomics techniques into patient and healthcare. These benefits include short-term and long-term benefits. Multiomics approaches have provided a large number of potential biomarkers and targets, which have produced short-term benefits with clear examples described above. Nevertheless, it will take a long time to fulfill the long-term benefits such as sensitive early diagnosis and significantly improved overall survival.
Multiomics technologies have generated an enormous amount of information critical to expanding our understanding of cancer biology and benefited the treatment of tumor patients. For example, in addition to analyzing tissue biopsy, whole genome sequencing could also be used in the circulation of cancer patients. Several studies have demonstrated the ability of whole genome sequencing in detecting chromosomal copy number changes, rearrangements, DNA hypomethylation, SNP and tumor heterogeneity [291,292,293]. This approach represents a useful method for noninvasive dynamic detection and monitoring of human tumors that is not dependent on the availability of tumor biopsies, which will bring benefits to patients who do not fit to biopsy. NGS benefits greatly to patients with rare cancers and cancer of unknown primary site, for detailed genomic profiling could be used to identify the main drivers of malignant transformation and to cover the shortage of diagnosis and treatment strategies [294, 295]. Linking genomic and proteomic data for biomarker and therapeutic target at the protein levels accelerate the drug development and benefit special subgroups of cancer patients [296]. Recent years, many novel targeted drugs have been developed and their clinical outcomes have been evaluated. Imatinib mesylate is highly efficacious in chronic myeloid leukemias and gastrointestinal stromal tumors [297, 298]. Non-squamous NSCLC patients with EGFR mutation benefited from gefitinib and afatinib with increased tumor response rate and prolonged progression-free survival compared to cytotoxic chemotherapy [299], while sorafenib may derive clinical benefit to NSCLC patients with wild-type EGFR [300]. Although a series of potential biomarkers generated by proteomics, metabolomics, and radiomics have not been approved in the clinical application, some of these candidates (such as AFP-L3 and des-γ-carboxyprothrombin in HCC [156, 301, 302], and sarcosine in Pca [232]) show better sensitivity and specificity compared to the FDA-approved biomarkers. More cancer patients will benefit from these biomarkers, if these biomarkers be validated in follow-up studies.
Conclusions and expert recommendations
The development of high-throughput and cost-effective multiple omics technologies have extensively used in in-depth understanding of the initiation, progression, and efficacious treatment of a cancer. DNA sequencing technologies, especially the NGS technologies, can detect a more comprehensive character of each major alternation in cancer genome. RNA-seq is a powerful tool to analyze gene expression profiles, and discovers novel intragenic fusion, somatic nucleotide mutations, transcripts, alternative splice forms, and non-coding RNAs. This genome profiling has the potential role in establishing different molecular subtypes and stratification of different patients, which is crucial in precisely personalized treatment. DNA and RNA are vectors of genetic information, and could reflect what will happen in the cells. Proteins encoded by the genes are ultimately the functional performer and could reflect what is really happening in real time or has happen in a given condition. MS-based proteomics demonstrate the powerful role in discovery of new biomarkers, driver events, and personalized therapeutic target, with access to a wide range of protein information from tissues and body fluids of cancer patients. Metabolomics not only provides results from complex gene-environment interactions under any conditions but also can feedback information on physiological and pathological processes. NMR- and MS-based metabolomics can effectively address scientific problems of a cancer, and have made obviously achievements in cancer diagnosis, assessment of response to traditional therapy, and discovery of novel drugs and therapeutics. Radiomics is the bridge between medical imaging and personalized medicine and could objectively and precisely provide detailed quantitative features of intratumoural and intertumoural heterogeneity in a non-invasive manner. Moreover, cancer is essentially a complex disease. Integrative multi-omics data provide a holistic view of the complexity in tumorigenesis, and benefit selection of right patients for targeted therapies and evaluation of traditional treatment strategies for improvement of its therapeutic effects. The multi-omics technologies have make significant achievements in cancer research and clinically relevant outcomes, and will surely accelerate the cancer research with the breakthrough of technical limitations and ultimately benefit more cancer patients in the world.
We recommend this review article to promote the education program regarding the roles of multi-omics in cancer research and clinically relevant outcomes, and emphasize the scientific importance of multi-omics in PPPM in a cancer, especially in discovery of multi-omics-based biomarkers for predictive diagnosis and prognosis assessment of a cancer, and in systematical clarification of molecular mechanisms to discover effectively therapeutic targets for a cancer.
Abbreviations
- CAGE:
-
Cap analysis of gene expression
- CID:
-
Collision induced dissociation
- CML:
-
Chronic myelogenous leukemia
- ECD:
-
Electro capture dissociation
- ETD:
-
Electron transfer dissociation
- FTICR:
-
Fourier transform ion cyclotron resonance
- GC:
-
Gas chromatography
- HCC:
-
Hepatocellular carcinoma
- HP:
-
Fucosylated haptoglobin
- HPLC:
-
High performance liquid chromatography
- IARC:
-
International Agency for Research on Cancer
- ICGC:
-
International Cancer Genome Consortium
- lncRNAs:
-
Long ncRNAs
- LC:
-
Liquid chromatography
- LSCC:
-
Lung squamous cell carcinoma
- MALDI:
-
Matrix-assisted laser desorption ionization
- MDLC:
-
Multi-dimensional LC
- MPSS:
-
Massively parallel signature sequencing
- MRM:
-
Multiple reaction monitoring
- MS:
-
Mass spectrometry
- MS/MS:
-
Tandem mass spectrometry
- NANA:
-
N-acetylneuraminic acid
- ncRNAs:
-
Non-coding RNAs
- NGS:
-
Next-generation sequencing
- NMR:
-
Nuclear magnetic resonance
- NSCLC:
-
Non-small cell lung cancer
- PPPM:
-
Predictive, preventive, and personalized medicine
- SAGE:
-
Serial analysis of gene expression
- SCLC:
-
Small cell lung cancer
- SCO:
-
Small cell osteosarcoma
- SRM:
-
Selected reaction monitoring
- SWATH:
-
Sequential window acquisition of all theoretical mass spectra
- TCGA:
-
Cancer Genome Atlas
- TOF:
-
Time-of-flight
- 1DGE:
-
One-dimensional gel electrophoresis
- 2DGE:
-
Two-dimensional gel electrophoresis
- 2D-DIGE:
-
Two-dimensional difference in-gel electrophoresis
- WGS:
-
Whole genome sequencing
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Tran B, Dancey JE, Kamel-Reid S, McPherson JD, Bedard PL, Brown AM, et al. Cancer genomics: technology, discovery, and translation. J Clin Oncol. 2012;30:647–60.
Horgan RP, Kenny LC. ‘Omic’ technologies: genomics, transcriptomics, proteomics and metabolomics. Obstet Gynaecol. 2011;13:189–95.
Chmielecki J, Meyerson M. DNA sequencing of cancer: what have we learned? Annu Rev Med. 2014;65:63–79.
Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9.
Vignot S, Frampton GM, Soria JC, Yelensky R, Commo F, Brambilla C, et al. Next-generation sequencing reveals high concordance of recurrent somatic alterations between primary tumor and metastases from patients with non-small-cell lung cancer. J Clin Oncol. 2013;31:2167–72.
Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–9.
Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–500.
Parsons DW, Jones S, Zhang X, Lin JC-H, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12.
Schuster SC. Next-generation sequencing transforms today's biology. Nat Methods. 2008;5:16.
Zhang H, Chan DW. Cancer biomarker discovery in plasma using a tissue-targeted proteomic approach. Cancer Epidemiol Biomark Prev. 2007;16:1915–7.
Sleno L, Emili A. Proteomic methods for drug target discovery. Curr Opin Chem Biol. 2008;12:46–54.
Johann DJ Jr, Wei BR, Prieto DA, Chan KC, Ye X, Valera VA, et al. Combined blood/tissue analysis for cancer biomarker discovery: application to renal cell carcinoma. Anal Chem. 2010;82:1584–8.
Ganti S, Taylor SL, Abu Aboud O, Yang J, Evans C, Osier MV, et al. Kidney tumor biomarkers revealed by simultaneous multiple matrix metabolomics analysis. Cancer Res. 2012;72:3471–9.
Alessandro R, Belluco C, Kohn EC. Proteomic approaches in colon cancer: promising tools for new cancer markers and drug target discovery. Clin Colorectal Cancer. 2005;4:396–402.
Zhang Z, Bast RC, Yu Y, Li J, Sokoll LJ, Rai AJ, et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882–90.
Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–304.
Hu R, Wang X, Zhan X. Multi-parameter systematic strategies for predictive, preventive and personalised medicine in cancer. EPMA J. 2013;4:2.
Tian Q, Price ND, Hood L. Systems cancer medicine: towards realization of predictive, preventive, personalized and participatory (P4) medicine. J Intern Med. 2012;271:111–21.
Hornberg JJ, Bruggeman FJ, Westerhoff HV, Lankelma J. Cancer: a systems biology disease. Biosystems. 2006;83:81–90.
Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8:51-60.
Grech G, Zhan X, Yoo BC, Bubnov R, Hagan S, Danesi R, et al. EPMA position paper in cancer: current overview and future perspectives. EPMA J. 2015;6:9.
Golubnitschaja O, Costigliola V. General report & recommendations in predictive, preventive and personalised medicine 2012: white paper of the European Association for Predictive, Preventive and Personalised Medicine. EPMA J. 2012;3:14.
Golubnitschaja O, Baban B, Boniolo G, Wang W, Bubnov R, Kapalla M, et al. Medicine in the early twenty-first century: paradigm and anticipation-EPMA position paper 2016. EPMA J. 2016;7:23.
Calkins GN. Zur frage der entstehung maligner tumoren. Science. 1914;40:857–9.
Rowley JD. A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–3.
Reddy EP, Reynolds RK, Santos E, Barbacid M. A point mutation is responsible for the acquisition of transforming properties by the T 24 human bladder carcinoma oncogene. Nature. 1982;300:149–52.
Macaluso M, Russo G, Cinti C, Bazan V, Gebbia N, Russo A. Ras family genes: an interesting link between cell cycle and cancer. J Cell Physiol. 2002;192:125–30.
Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–62.
Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, et al. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099–109.
Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–96.
Collins FS, Barker AD. Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am. 2007;296:50–7.
Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci. 1977;74:5463–7.
Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94:441IN19447–6IN20448.
Sanger F. Determination of nucleotide sequences in DNA. Biosci Rep. 1981;1:3–18.
Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46.
Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–45.
Ansorge WJ. Next-generation DNA sequencing techniques. New Biotechnol. 2009;25:195–203.
Ajay SS, Parker SC, Abaan HO, Fajardo KVF, Margulies EH. Accurate and comprehensive sequencing of personal genomes. Genome Res. 2011;21:1498–505.
Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–64.
Niedringhaus TP, Milanova D, Kerby MB, Snyder MP, Barron AE. Landscape of next-generation sequencing technologies. Anal Chem. 2011;83:4327.
Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–24.
Network CGAR. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519.
Bass AJ, Thorsson V, Shmulevich I, Reynolds SM, Miller M, Bernard B, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202.
Network CGA. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330.
Network CGAR. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061.
Network CGAR. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609.
Marchetti A, Del Grammastro M, Felicioni L, Malatesta S, Filice G, Centi I, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One. 2014;9:e103883.
Hagemann IS, Devarakonda S, Lockwood CM, Spencer DH, Guebert K, Bredemeyer AJ, et al. Clinical next-generation sequencing in patients with non–small cell lung cancer. Cancer. 2015;121:631–9.
Beltran H, Yelensky R, Frampton GM, Park K, Downing SR, MacDonald TY, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–6.
Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214.
Weisman PS, Ng CK, Brogi E, Eisenberg RE, Won HH, Piscuoglio S, et al. Genetic alterations of triple negative breast cancer by targeted next generation sequencing and correlation with tumor morphology. Mod Pathol. 2016;29:476.
Janku F, Kaseb AO, Tsimberidou AM, Wolff RA, Kurzrock R. Identification of novel therapeutic targets in the PI3K/AKT/mTOR pathway in hepatocellular carcinoma using targeted next generation sequencing. Oncotarget. 2014;5:3012.
Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, Jones DM, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist. 2014;19:235–42.
Ward DG, Baxter L, Gordon NS, Ott S, Savage RS, Beggs AD, et al. Multiplex PCR and next generation sequencing for the non-invasive detection of bladder cancer. PLoS One. 2016;11:e0149756.
Liang WS, Craig DW, Carpten J, Borad MJ, Demeure MJ, Weiss GJ, et al. Genome-wide characterization of pancreatic adenocarcinoma patients using next generation sequencing. PLoS One. 2012;7:e43192.
Kim PH, Cha EK, Sfakianos JP, Iyer G, Zabor EC, Scott SN, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol. 2015;67:198–201.
Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45.
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8.
Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6.
Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–76.
Palanisamy N, Ateeq B, Kalyana-Sundaram S, Pflueger D, Ramnarayanan K, Shankar S, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16:793–8.
Ross JS, Wang K, Chmielecki J, Gay L, Johnson A, Chudnovsky J, et al. The distribution of BRAF gene fusions in solid tumors and response to targeted therapy. Int J Cancer. 2016;138:881–90.
Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM. Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med. 2015;7:129.
Tomlins SA, Day JR, Lonigro RJ, Hovelson DH, Siddiqui J, Kunju LP, et al. Urine TMPRSS2: ERG plus PCA3 for individualized prostate cancer risk assessment. Eur Urol. 2016;70:45–53.
Leyten GH, Hessels D, Jannink SA, Smit FP, de Jong H, Cornel EB, et al. Prospective multicentre evaluation of PCA3 and TMPRSS2-ERG gene fusions as diagnostic and prognostic urinary biomarkers for prostate cancer. Eur Urol. 2014;65:534–42.
Salagierski M, Schalken JA. Molecular diagnosis of prostate cancer: PCA3 and TMPRSS2: ERG gene fusion. J Urol. 2012;187:795–801.
Skálová A, Vanecek T, Sima R, Laco J, Weinreb I, Perez-Ordonez B, et al. Mammary analogue secretory carcinoma of salivary glands, containing the ETV6-NTRK3 fusion gene: a hitherto undescribed salivary gland tumor entity. Am J Surg Pathol. 2010;34:599–608.
Morrison KB, Tognon CE, Garnett MJ, Deal C, Sorensen PH. ETV6-NTRK3 transformation requires insulin-like growth factor 1 receptor signaling and is associated with constitutive IRS-1 tyrosine phosphorylation. Oncogene. 2002;21:5684.
Tognon C, Garnett M, Kenward E, Kay R, Morrison K, Sorensen PH. The chimeric protein tyrosine kinase ETV6-NTRK3 requires both Ras-Erk1/2 and PI3-kinase-Akt signaling for fibroblast transformation. Cancer Res. 2001;61:8909–16.
Khotskaya YB, Holla VR, Farago AF, Shaw KRM, Meric-Bernstam F, Hong DS. Targeting TRK family proteins in cancer. Pharmacol Ther. 2017;173:58-6.
Capelletti M, Dodge ME, Ercan D, Hammerman PS, Park S-I, Kim J, et al. Identification of recurrent FGFR3–TACC3 fusion oncogenes from lung adenocarcinoma. Clin Cancer Res. 2014;20:6551–8.
Okamoto I, Nakagawa K. Echinoderm microtubule-associated protein-like 4–anaplastic lymphoma kinase-targeted therapy for advanced non-small cell lung cancer: molecular and clinical aspects. Cancer Sci. 2012;103:1391–6.
Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63.
Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE Jr, et al. Characterization of the yeast transcriptome. Cell. 1997;88:243–51.
Duggan DJ, Bittner M, Chen Y, Meltzer P, Trent JM. Expression profiling using cDNA microarrays. Nat Genet. 1999;21:10–4.
Yazaki J, Gregory BD, Ecker JR. Mapping the genome landscape using tiling array technology. Curr Opin Plant Biol. 2007;10:534–42.
Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457-63.
Wang G-S, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–61.
Liu S, Cheng C. Alternative RNA splicing and cancer. Wiley Interdiscip Rev RNA. 2013;4:547–66.
Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–54.
Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z, et al. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett. 2017;393:40–51.
Frasca F, Pandini G, Scalia P, Sciacca L, Mineo R, Costantino A, et al. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol Cell Biol. 1999;19:3278–88.
Jiang L, Zhu W, Streicher K, Morehouse C, Brohawn P, Ge X, et al. Increased IR-A/IR-B ratio in non-small cell lung cancers associates with lower epithelial-mesenchymal transition signature and longer survival in squamous cell lung carcinoma. BMC Cancer. 2014;14:131.
Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, et al. An EMT–driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011;7:e1002218.
Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101.
Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98.
Kekeeva T, Tanas A, Kanygina A, Alexeev D, Shikeeva A, Zavalishina L, et al. Novel fusion transcripts in bladder cancer identified by RNA-seq. Cancer Lett. 2016;374:224–8.
Kloosterman WP, van den Braak RRC, Pieterse M, van Roosmalen MJ, Sieuwerts AM, Stangl C, et al. A systematic analysis of oncogenic gene fusions in primary colon cancer. Cancer Res. 2017;77:3814–22.
McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, et al. deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol. 2011;7:e1001138.
Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13:358–69.
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8.
O'donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Law PT-Y, Qin H, Ching AK-K, Lai KP, Co NN, He M, et al. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58:1165–73.
Beck D, Ayers S, Wen J, Brandl MB, Pham TD, Webb P, et al. Integrative analysis of next generation sequencing for small non-coding RNAs and transcriptional regulation in Myelodysplastic Syndromes. BMC Med Genet. 2011;4:19.
Xie L, Liao Y, Shen L, Hu F, Yu S, Zhou Y, et al. Identification of the miRNA-mRNA regulatory network of small cell osteosarcoma based on RNA-seq. Oncotarget. 2017;8:42525-36.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6.
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–19.
Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26:155–65.
Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21.
Yang Z, Zhou L, Wu L-M, Lai M-C, Xie H-Y, Zhang F, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18:1243–50.
M-c L, Yang Z, Zhou L, Zhu Q-Q, Xie H-Y, Zhang F, et al. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–6.
Gutschner T, Hämmerle M, Eißmann M, Hsu J, Kim Y, Hung G, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–9.
Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9.
Zhang L, Li S, Choi Y-L, Lee J, Gong Z, Liu X, et al. Systematic identification of cancer-related long noncoding RNAs and aberrant alternative splicing of quintuple-negative lung adenocarcinoma through RNA-Seq. Lung Cancer. 2017;109:21–7.
Tang R, Chen W, He R, Zeng J, Liang L, Li S, et al. Identification of a RNA-Seq based prognostic signature with five lncRNAs for lung squamous cell carcinoma. Oncotarget. 2017;8:50761-73
Fan Q, Liu B. Identification of a RNA-Seq based 8-long non-coding RNA signature predicting survival in esophageal cancer. Med Sci Monit. 2016;22:5163.
Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–8.
Stastna M, Van Eyk JE. Analysis of protein isoforms: can we do it better? Proteomics. 2012;12:2937–48.
Zhan X, Giorgianni F, Desiderio DM. Proteomics analysis of growth hormone isoforms in the human pituitary. Proteomics. 2005;5:1228–41.
Zhan X, Long Y, Lu M. Exploration of variations in proteome and metabolome for predictive diagnostics and personalized treatment algorithms: innovative approach and examples for potential clinical application. J Proteomics. 2017. https://doi.org/10.1016/j.jprot.2017.08.020.
Zhan X, Yang H, Peng F, Li J, Mu Y, Long Y, et al. How many proteins can be identified in a 2-DE gel spot within an analysis of a complex human cancer tissue proteome? Electrophoresis. 2017; https://doi.org/10.1002/elps.201700330.
Kohler M, Thomas A, Püschel K, Schänzer W, Thevis M. Identification of human pituitary growth hormone variants by mass spectrometry. J Proteome Res. 2009;8:1071–6.
Peng F, Li J, Guo T, Yang H, Li M, Sang S, et al. Nitroproteins in human astrocytomas discovered by gel electrophoresis and tandem mass spectrometry. J Am Soc Mass Spectrom. 2015;26:2062–76.
Zhan X, Desiderio DM. The human pituitary nitroproteome: detection of nitrotyrosyl-proteins with two-dimensional Western blotting, and amino acid sequence determination with mass spectrometry. Biochem Biophys Res Commun. 2004;325:1180–6.
Guo T, Wang X, Li M, Yang H, Li L, Peng F, et al. Identification of glioblastoma phosphotyrosine-containing proteins with two-dimensional western blotting and tandem mass spectrometry. Biomed Res Int. 2015;2015:134050.
Goheen SC, Engelhorn SC. Hydrophobic interaction high-performance liquid chromatography of proteins. J Chromatogr A. 1984;317:55–65.
Staub A, Zurlino D, Rudaz S, Veuthey J-L, Guillarme D. Analysis of peptides and proteins using sub-2μm fully porous and sub 3-μm shell particles. J Chromatogr A. 2011;1218:8903–14.
Geng X, Ke C, Chen G, Liu P, Wang F, Zhang H, et al. On-line separation of native proteins by two-dimensional liquid chromatography using a single column. J Chromatogr A. 2009;1216:3553–62.
Sikanen T, Aura S, Franssila S, Kotiaho T, Kostiainen R. Microchip capillary electrophoresis–electrospray ionization–mass spectrometry of intact proteins using uncoated Ormocomp microchips. Anal Chim Acta. 2012;711:69–76.
Tran JC, Doucette AA. Multiplexed size separation of intact proteins in solution phase for mass spectrometry. Anal Chem. 2009;81:6201–9.
Fagerquist CK, Sultan O. Induction and identification of disulfide-intact and disulfide-reduced β-subunit of Shiga toxin 2 from Escherichia coli O157: H7 using MALDI-TOF-TOF-MS/MS and top-down proteomics. Analyst. 2011;136:1739–46.
Mao Y, Valeja SG, Rouse JC, Hendrickson CL, Marshall AG. Top-down structural analysis of an intact monoclonal antibody by electron capture dissociation-Fourier transform ion cyclotron resonance-mass spectrometry. Anal Chem. 2013;85:4239–46.
Tipton JD, Tran JC, Catherman AD, Ahlf DR, Durbin KR, Lee JE, et al. Nano-LC FTICR tandem mass spectrometry for top-down proteomics: routine baseline unit mass resolution of whole cell lysate proteins up to 72 kDa. Anal Chem. 2012;84:2111–7.
Tveen-Jensen K, Reis A, Spickett CM, Pitt AR. P93-Targeted mass spectrometry methods for detecting oxidative post-translational modifications. Free Radic Biol Med. 2014;75:S52–S3.
Scheffler K. Top-down proteomics by means of Orbitrap mass spectrometry. Methods Mol Biol. 2014;1156:465–87.
Brunner AM, Lössl P, Liu F, Huguet R, Mullen C, Yamashita M, et al. Benchmarking multiple fragmentation methods on an orbitrap fusion for top-down phospho-proteoform characterization. Anal Chem. 2015;87:4152–8.
Takayama M, Sekiya S, Iimuro R, Iwamoto S, Tanaka K. Selective and nonselective cleavages in positive and negative CID of the fragments generated from in-source decay of intact proteins in MALDI-MS. J Am Soc Mass Spectrom. 2014;25:120–31.
Riley NM, Westphall MS, Coon JJ. Activated ion electron transfer dissociation for improved fragmentation of intact proteins. Anal Chem. 2015;87:7109–16.
Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Native electrospray and electron-capture dissociation in FTICR mass spectrometry provide top-down sequencing of a protein component in an intact protein assembly. J Am Soc Mass Spectrom. 2010;21:1966–8.
Mn C, Cañas B, Js V, Gallardo JM. Extensive de novo sequencing of new parvalbumin isoforms using a novel combination of bottom-up proteomics, accurate molecular mass measurement by FTICR− MS, and selected MS/MS Ion monitoring. J Proteome Res. 2010;9:4393–406.
Durbin KR, Fornelli L, Fellers RT, Doubleday PF, Narita M, Kelleher NL. Quantitation and identification of thousands of human proteoforms below 30 kDa. J Proteome Res. 2016;15:976–82.
Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207.
Zhang Y, Fonslow BR, Shan B, Baek M-C, Yates JR III. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113:2343–94.
Arentz G, Weiland F, Oehler MK, Hoffmann P. State of the art of 2D DIGE. Proteomics Clin Appl. 2015;9:277–88.
Collier TS, Muddiman DC. Analytical strategies for the global quantification of intact proteins. Amino Acids. 2012;43:1109-17.
Nie S, Lo A, Zhu J, Wu J, Ruffin MT, Lubman DM. Isobaric protein-level labeling strategy for serum glycoprotein quantification analysis by liquid chromatography–tandem mass spectrometry. Anal Chem. 2013;85:5353–7.
Karabudak A, Hafner J, Shetty V, Chen S, Secord AA, Morse M, et al. Autoantibody biomarkers identified by proteomics methods distinguish ovarian cancer from non ovarian cancer with various CA-125 levels. J Cancer Res Clin Oncol. 2013;139:1757–70.
Merl J, Deeg CA, Swadzba ME, Ueffing M, Hauck SM. Identification of autoantigens in body fluids by combining pull-downs and organic precipitations of intact immune complexes with quantitative label-free mass spectrometry. J Proteome Res. 2013;12:5656–65.
Russell JD, Scalf M, Book AJ, Ladror DT, Vierstra RD, Smith LM, et al. Characterization and quantification of intact 26S proteasome proteins by real-time measurement of intrinsic fluorescence prior to top-down mass spectrometry. PLoS One. 2013;8:e58157.
Oeckl P, Steinacker P, von Arnim CA, Straub S, Nagl M, Feneberg E, et al. Intact protein analysis of ubiquitin in cerebrospinal fluid by multiple reaction monitoring reveals differences in Alzheimer’s disease and frontotemporal lobar degeneration. J Proteome Res. 2014;13:4518–25.
Janecki DJ, Bemis KG, Tegeler TJ, Sanghani PC, Zhai L, Hurley TD, et al. A multiple reaction monitoring method for absolute quantification of the human liver alcohol dehydrogenase ADH1C1 isoenzyme. Anal Biochem. 2007;369:18–26.
Sidoli S, Lin S, Xiong L, Bhanu NV, Karch KR, Johansen E, et al. Sequential window acquisition of all theoretical mass spectra (SWATH) analysis for characterization and quantification of histone post-translational modifications. Mol Cell Proteomics. 2015;14:2420–8.
Collins BC, Gillet LC, Rosenberger G, Röst HL, Vichalkovski A, Gstaiger M, et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat Methods. 2013;10:1246–53.
Hyung SJ, Ruotolo BT. Integrating mass spectrometry of intact protein complexes into structural proteomics. Proteomics. 2012;12:1547–64.
Zhan X, Wang X, Desiderio DM. Mass spectrometry analysis of nitrotyrosine-containing proteins. Mass Spectrom Rev. 2015;34:423–48.
Hägglund P, Bunkenborg J, Elortza F, Jensen ON, Roepstorff P. A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J Proteome Res. 2004;3:556–66.
Ahn J-M, Sung H-J, Yoon Y-H, Kim B-G, Yang WS, Lee C, et al. Integrated glycoproteomics demonstrates fucosylated serum paraoxonase 1 alterations in small cell lung cancer. Mol Cell Proteomics. 2014;13:30–48.
Tsai HY, Boonyapranai K, Sriyam S, Yu CJ, Wu SW, Khoo KH, et al. Glycoproteomics analysis to identify a glycoform on haptoglobin associated with lung cancer. Proteomics. 2011;11:2162–70.
Liu Y, He J, Li C, Benitez R, Fu S, Marrero J, et al. Identification and confirmation of biomarkers using an integrated platform for quantitative analysis of glycoproteins and their glycosylations. J Proteome Res. 2009;9:798–805.
Li D, Mallory T, Satomura S. AFP-L3: a new generation of tumor marker for hepatocellular carcinoma. Clin Chim Acta. 2001;313:15–9.
Andriole GL, Crawford ED, Grubb RL III, Buys SS, Chia D, Church TR, et al. Mortality results from a randomized prostate-cancer screening trial. N Engl J Med. 2009;360:1310–9.
Health NIo. Role of active surveillance in the management of men with localized prostate cancer. State-of-the-Science Conference Draft Statement. 2011;28:1-27.
Ip S, Dahabreh IJ, Chung M, Yu WW, Balk EM, Iovin RC, et al. An evidence review of active surveillance in men with localized prostate cancer. Evid Rep Technol Assess. 2011;204:1-341.
Liu Y, Chen J, Sethi A, Li QK, Chen L, Collins B, et al. Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers N-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Mol Cell Proteomics. 2014;13:1753–68.
Zawadzka AM, Schilling B, Cusack MP, Sahu AK, Drake P, Fisher SJ, et al. Phosphoprotein secretome of tumor cells as a source of candidates for breast cancer biomarkers in plasma. Mol Cell Proteomics. 2014;13:1034–49.
Craven RJ. PGRMC1: a new biomarker for the estrogen receptor in breast cancer. Breast Cancer Res. 2008;10:113.
Gámez-Pozo A, Sánchez-Navarro I, Calvo E, Agulló-Ortuño MT, López-Vacas R, Díaz E, et al. PTRF/cavin-1 and MIF proteins are identified as non-small cell lung cancer biomarkers by label-free proteomics. PLoS One. 2012;7:e33752.
Zhan X, Wang X, Desiderio DM. Pituitary adenoma nitroproteomics: current status and perspectives. Oxidative Med Cell Longev. 2013;2013:580710.
Guo T, Zhu Y, Gan CS, Lee SS, Zhu J, Wang H, et al. Quantitative proteomics discloses MET expression in mitochondria as a direct target of MET kinase inhibitor in cancer cells. Mol Cell Proteomics. 2010;9:2629–41.
Van Houdt WJ, Emmink BL, Pham TV, Piersma SR, Verheem A, Vries R, et al. Comparative proteomics of colon cancer stem cells and differentiated tumor cells identifies BIRC6 as a potential therapeutic target. Mol Cell Proteomics. 2011;10:M111-011353.
Yokoyama T, Enomoto T, Serada S, Morimoto A, Matsuzaki S, Ueda Y, et al. Plasma membrane proteomics identifies bone marrow stromal antigen 2 as a potential therapeutic target in endometrial cancer. Int J Cancer. 2013;132:472–84.
Li Z, Zhao X, Bai S, Wang Z, Chen L, Wei Y, et al. Proteomics identification of cyclophilin a as a potential prognostic factor and therapeutic target in endometrial carcinoma. Mol Cell Proteomics. 2008;7:1810–23.
Ren F, Wu H, Lei Y, Zhang H, Liu R, Zhao Y, et al. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol Cancer. 2010;9:81.
Ren H, Tan Z-P, Zhu X, Crosby K, Haack H, Ren J-M, et al. Identification of anaplastic lymphoma kinase as a potential therapeutic target in ovarian cancer. Cancer Res. 2012;72:3312–23.
Balabanov S, Gontarewicz A, Ziegler P, Hartmann U, Kammer W, Copland M, et al. Hypusination of eukaryotic initiation factor 5A (eIF5A): a novel therapeutic target in BCR-ABL–positive leukemias identified by a proteomics approach. Blood. 2007;109:1701–11.
Zhan X, Desiderio DM. Signaling pathway networks mined from human pituitary adenoma proteomics data. BMC Med Genet. 2010;3:13.
Qin G, Dang M, Gao H, Wang H, Luo F, Chen R. Deciphering the protein–protein interaction network regulating hepatocellular carcinoma metastasis. Biochim Biophys Acta (BBA)-Proteins Proteomics. 2017;1865:1114-22.
McKnight SL. On getting there from here. Science. 2010;330:1338–9.
Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134:714–7.
Patti GJ, Yanes O, Siuzdak G. Innovation: metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–9.
Nicholson JK. Global systems biology, personalized medicine and molecular epidemiology. Mol Syst Biol. 2006;2:52.
Mirsaeidi M, Banoei MM, Winston BW, Schraufnagel DE. Metabolomics: applications and promise in Mycobacterial disease. Ann Am Thorac Soc. 2015;12:1278–87.
Daye D, Wellen KE. Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin Cell Dev Biol. 2012;23:362–9.
Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem Biol. 2015;10:95–108.
Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, et al. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–5.
Bryant KL, Mancias JD, Kimmelman AC, Der CJ. KRAS: feeding pancreatic cancer proliferation. Trends Biochem Sci. 2014;39:91–100.
Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, Chan Q, et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400.
Serkova NJ, Glunde K. Metabolomics of cancer. Methods Mol Biol. 2009;520:273–95.
Tebani A, Abily-Donval L, Afonso C, Marret S, Bekri S. Clinical metabolomics: the new metabolic window for inborn errors of metabolism investigations in the post-genomic era. Int J Mol Sci. 2016;17:1167.
Spratlin JL, Serkova NJ, Eckhardt SG. Clinical applications of metabolomics in oncology: a review. Clin Cancer Res. 2009;15:431–40.
Keun HC, Ebbels TM, Antti H, Bollard ME, Beckonert O, Schlotterbeck G, et al. Analytical reproducibility in 1H NMR-based metabonomic urinalysis. Chem Res Toxicol. 2002;15:1380–6.
Dumas M-E, Maibaum EC, Teague C, Ueshima H, Zhou B, Lindon JC, et al. Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: the INTERMAP Study. Anal Chem. 2006;78:2199–208.
Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–703.
Sager JE, Choiniere JR, Chang J, Stephenson-Famy A, Nelson WL, Isoherranen N. Identification and structural characterization of three new metabolites of bupropion in humans. ACS Med Chem Lett. 2016;7:791–6.
Halabalaki M, Vougogiannopoulou K, Mikros E, Skaltsounis AL. Recent advances and new strategies in the NMR-based identification of natural products. Curr Opin Biotechnol. 2014;25:1–7.
Li C, Lee M-J, Sheng S, Meng X, Prabhu S, Winnik B, et al. Structural identification of two metabolites of catechins and their kinetics in human urine and blood after tea ingestion. Chem Res Toxicol. 2000;13:177–84.
Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TM, Holmes E, et al. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc. 2010;5:1019–32.
Jordan KW, Nordenstam J, Lauwers GY, Rothenberger DA, Alavi K, Garwood M, et al. Metabolomic characterization of human rectal adenocarcinoma with intact tissue magnetic resonance spectroscopy. Dis Colon Rectum. 2009;52:520.
Mirnezami R, Jiménez B, Li JV, Kinross JM, Veselkov K, Goldin RD, et al. Rapid diagnosis and staging of colorectal cancer via high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy of intact tissue biopsies. Ann Surg. 2014;259:1138–49.
Giskeødegård GF, Cao MD, Bathen TF. High-resolution magic-angle-spinning NMR spectroscopy of intact tissue. Methods Mol Biol. 2015;1277:37–50.
Fan TW, Lane AN. NMR-based stable isotope resolved metabolomics in systems biochemistry. J Biomol NMR. 2011;49:267–80.
Barding GA, Salditos R, Larive CK. Quantitative NMR for bioanalysis and metabolomics. Anal Bioanal Chem. 2012;404:1165–79.
Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal Bioanal Chem. 2007;387:525–7.
Fernie AR, Trethewey RN, Krotzky AJ, Willmitzer L. Metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol. 2004;5:763–9.
Bingol K, Brüschweiler R. NMR/MS translator for the enhanced simultaneous analysis of metabolomics mixtures by NMR spectroscopy and mass spectrometry: application to human urine. J Proteome Res. 2015;14:2642–8.
Madhu B, Shaw GL, Warren AY, Neal DE, Griffiths JR. Response of Degarelix treatment in human prostate cancer monitored by HR-MAS 1. Metabolomics. 2016;12:1–11.
Hajduk A, Mrochem-Kwarciak J, Skorupa A, Ciszek M, Heyda A, Składowski K, et al. 1H NMR based metabolomic approach to monitoring of the head and neck cancer treatment toxicity. Metabolomics. 2016;12:1–15.
Hadi NI, Jamal Q, Iqbal A, Shaikh F, Somroo S, Musharraf SG. Serum metabolomic profiles for breast cancer diagnosis, grading and staging by gas chromatography-mass spectrometry. Sci Rep. 2017;7:1715.
Cameron SJ, Lewis KE, Beckmann M, Allison GG, Ghosal R, Lewis PD, et al. The metabolomic detection of lung cancer biomarkers in sputum. Lung Cancer. 2016;94:88–95.
Di Gangi IM, Mazza T, Fontana A, Copetti M, Fusilli C, Ippolito A, et al. Metabolomic profile in pancreatic cancer patients: a consensus-based approach to identify highly discriminating metabolites. Oncotarget. 2016;7:5815.
Hou Y, Yin M, Sun F, Zhang T, Zhou X, Li H, et al. A metabolomics approach for predicting the response to neoadjuvant chemotherapy in cervical cancer patients. Mol BioSyst. 2014;10:2126–33.
Mathé EA, Patterson AD, Haznadar M, Manna SK, Krausz KW, Bowman ED, et al. Noninvasive urinary metabolomic profiling identifies diagnostic and prognostic markers in lung cancer. Cancer Res. 2014;74:3259–70.
Armitage EG, Barbas C. Metabolomics in cancer biomarker discovery: current trends and future perspectives. J Pharm Biomed Anal. 2014;87:1–11.
Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–52.
Calderón-Santiago M, Priego-Capote F, Turck N, Robin X, Jurado-Gámez B, Sanchez JC, et al. Human sweat metabolomics for lung cancer screening. Anal Bioanal Chem. 2015;407:5381–92.
Takayama T, Tsutsui H, Shimizu I, Toyama T, Yoshimoto N, Endo Y, et al. Diagnostic approach to breast cancer patients based on target metabolomics in saliva by liquid chromatography with tandem mass spectrometry. Clin Chim Acta. 2016;452:18–26.
Kelly RS, Vander Heiden MG, Giovannucci E, Mucci LA. Metabolomic biomarkers of prostate cancer: prediction, diagnosis, progression, prognosis, and recurrence. Cancer Epidemiol Prev Biomark. 2016;25:887-906.
Zhang T, Watson DG, Wang L, Abbas M, Murdoch L, Bashford L, et al. Application of holistic liquid chromatography-high resolution mass spectrometry based urinary metabolomics for prostate cancer detection and biomarker discovery. PLoS One. 2013;8:e65880.
McDunn JE, Stirdivant SM, Ford LA, Wolfert RL. Metabolomics and its application to the development of clinical laboratory tests for prostate cancer. EJIFCC. 2015;26:92.
Jin X, Yun SJ, Jeong P, Kim IY, Kim W-J, Park S. Diagnosis of bladder cancer and prediction of survival by urinary metabolomics. Oncotarget. 2014;5:1635–45.
Wittmann BM, Stirdivant SM, Mitchell MW, Wulff JE, McDunn JE, Li Z, et al. Bladder cancer biomarker discovery using global metabolomic profiling of urine. PLoS One. 2014;9:e115870.
Peng J, Chen Y-T, Chen C-L, Li L. Development of a universal metabolome-standard method for long-term LC–MS metabolome profiling and its application for bladder cancer urine-metabolite-biomarker discovery. Anal Chem. 2014;86:6540–7.
Fan L, Yin M, Ke C, Ge T, Zhang G, Zhang W, et al. Use of plasma metabolomics to identify diagnostic biomarkers for early stage epithelial ovarian cancer. J Cancer. 2016;7:1265.
Turkoglu O, Zeb A, Graham S, Szyperski T, Szender JB, Odunsi K, et al. Metabolomics of biomarker discovery in ovarian cancer: a systematic review of the current literature. Metabolomics. 2016;12:1–16.
Tian Y, Bai H, Wang J, Wang J. PUB146 prognostic prediction of pemetrexed-platinum chemotherapeutic regimen by serum metabolomics. J Thorac Oncol. 2017;12:S1530–S1.
Di Gialleonardo V, Aldeborgh HN, Miloushev V, Folkers KM, Granlund K, Tap WD, et al. Multinuclear NMR and MRI reveal an early metabolic response to mTOR inhibition in sarcoma. Cancer Res. 2017;77:3113-20.
Seyfried TN, Shelton LM. Cancer as a metabolic disease. Nutr Metab. 2010;7:7.
Seyfried TN, Flores R, Poff AM, D’Agostino DP. Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis. 2013;35:515–27.
Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15:473-84.
Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–9.
Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010; https://doi.org/10.1084/jem.20092506jem.
Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders JE, Zeilemaker A, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010;116:2122–6.
Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406.
Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, George E, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54.
Khan AP, Rajendiran TM, Bushra A, Asangani IA, Athanikar JN, Yocum AK, et al. The role of sarcosine metabolism in prostate cancer progression. Neoplasia. 2013;15:491IN6–501IN13.
Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009;457:910–4.
Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–33.
Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123:3678–84.
Lorenzi PL, Reinhold WC, Rudelius M, Gunsior M, Shankavaram U, Bussey KJ, et al. Asparagine synthetase as a causal, predictive biomarker for L-asparaginase activity in ovarian cancer cells. Mol Cancer Ther. 2006;5:2613–23.
Gillies RJ, Gatenby RA. Metabolism and its sequelae in cancer evolution and therapy. Cancer J (Sudbury, Mass). 2015;21:88.
Hirschhaeuser F, Sattler UG, Mueller-Klieser W. Lactate: a metabolic key player in cancer. Cancer Res. 2011;71:6921–5.
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739.
Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34.
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30.
Chowdhury R, Yeoh KK, Tian YM, Hillringhaus L, Bagg EA, Rose NR, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–9.
Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–8.
Losman J-A, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–5.
Amary MF, Bacsi K, Maggiani F, Damato S, Halai D, Berisha F, et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J Pathol. 2011;224:334–43.
Lu C, Venneti S, Akalin A, Fang F, Ward PS, DeMatteo RG, et al. Induction of sarcomas by mutant IDH2. Genes Dev. 2013;27:1986–98.
Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, et al. Frequent mutation of isocitrate dehydrogenase (IDH) 1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–9.
Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–30.
Li L, Paz AC, Wilky BA, Johnson B, Galoian K, Rosenberg A, et al. Treatment with a small molecule mutant IDH1 inhibitor suppresses tumorigenic activity and decreases production of the oncometabolite 2-hydroxyglutarate in human chondrosarcoma cells. PLoS One. 2015;10:e0133813.
Wang F, Travins J, DeLaBarre B, Penard-Lacronique V, Schalm S, Hansen E, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–6.
DiNardo C, de Botton S, Pollyea DA, Stein EM, Fathi AT, Roboz GJ, et al. Molecular profiling and relationship with clinical response in patients with IDH1 mutation-positive hematologic malignancies receiving AG-120, a first-in-class potent inhibitor of mutant IDH1, in addition to data from the completed dose escalation portion of the phase 1 study. Am Soc Hematol. 2015;126:Abstract 1306.
Stein EM, Altman JK, Collins R, DeAngelo DJ, Fathi AT, Flinn I, et al. AG-221, an oral, selective, first-in-class, potent inhibitor of the IDH2 mutant metabolic enzyme, induces durable remissions in a phase I study in patients with IDH2 mutation positive advanced hematologic malignancies. Am Soc Hematol. 2014;124:Abstract 115.
Stein EM, DiNardo C, Altman JK, Collins R, DeAngelo DJ, Kantarjian HM, et al. Safety and efficacy of AG-221, a potent inhibitor of mutant IDH2 that promotes differentiation of myeloid cells in patients with advanced hematologic malignancies: results of a phase 1/2 trial. Am Soc Hematol. 2015;126:Abstract 323.
Dang L, Yen K, Attar E. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol. 2016;27:599–608.
Leszczyniecka M, Roberts T, Dent P, Grant S, Fisher PB. Differentiation therapy of human cancer: basic science and clinical applications. Pharmacol Ther. 2001;90:105–56.
Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–48.
Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–6.
Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278:563–77.
Lambin P, Leijenaar RT, Deist TM, Peerlings J, de Jong EE, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749.
Diehn M, Nardini C, Wang DS, McGovern S, Jayaraman M, Liang Y, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci. 2008;105:5213–8.
Yamamoto S, Korn RL, Oklu R, Migdal C, Gotway MB, Weiss GJ, et al. ALK molecular phenotype in non–small cell lung cancer: CT radiogenomic characterization. Radiology. 2014;272:568–76.
Fanchon LM, Dogan S, Moreira AL, Carlin SA, Schmidtlein CR, Yorke E, et al. Feasibility of in situ, high-resolution correlation of tracer uptake with histopathology by quantitative autoradiography of biopsy specimens obtained under 18F-FDG PET/CT guidance. J Nucl Med. 2015;56:538–44.
Parmar C, Velazquez ER, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One. 2014;9:e102107.
Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Cavalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006.
Cook GJ, Yip C, Siddique M, Goh V, Chicklore S, Roy A, et al. Are pretreatment 18F-FDG PET tumor textural features in non–small cell lung cancer associated with response and survival after chemoradiotherapy? J Nucl Med. 2013;54:19–26.
Coroller TP, Grossmann P, Hou Y, Velazquez ER, Leijenaar RT, Hermann G, et al. CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114:345–50.
Yang D, Rao G, Martinez J, Veeraraghavan A, Rao A. Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med Phys. 2015;42:6725–35.
McGarry SD, Hurrell SL, Kaczmarowski AL, Cochran EJ, Connelly J, Rand SD, et al. Magnetic resonance imaging-based radiomic profiles predict patient prognosis in newly diagnosed glioblastoma before therapy. Tomography. 2016;2:223.
Wibmer A, Hricak H, Gondo T, Matsumoto K, Veeraraghavan H, Fehr D, et al. Haralick texture analysis of prostate MRI: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different Gleason scores. Eur Radiol. 2015;25:2840–50.
Nketiah G, Elschot M, Kim E, Teruel JR, Scheenen TW, Bathen TF, et al. T2-weighted MRI-derived textural features reflect prostate cancer aggressiveness: preliminary results. Eur Radiol. 2017;27:3050–9.
Cunliffe A, Armato SG, Castillo R, Pham N, Guerrero T, Al-Hallaq HA. Lung texture in serial thoracic computed tomography scans: correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development. Int J Radiat Oncol Biol Phys. 2015;91:1048–56.
Tan S, Kligerman S, Chen W, Lu M, Kim G, Feigenberg S, et al. Spatial-temporal [18 F] FDG-PET features for predicting pathologic response of esophageal cancer to neoadjuvant chemoradiation therapy. Int J Radiat Oncol Biol Phys. 2013;85:1375–82.
Nie K, Shi L, Chen Q, Hu X, Jabbour SK, Yue N, et al. Rectal cancer: assessment of neoadjuvant chemoradiation outcome based on radiomics of multiparametric MRI. Clin Cancer Res. 2016;22:5256–64.
Obeid J-P, Stoyanova R, Kwon D, Patel M, Padgett K, Slingerland J, et al. Multiparametric evaluation of preoperative MRI in early stage breast cancer: prognostic impact of peri-tumoral fat. Clin Transl Oncol. 2017;19:211–8.
Chen X, Bergom C, Currey A, Kelly T, Edwin C, Montes A, et al. Quantitative computed tomography for radiation-induced changes in normal breast tissue during partial breast irradiation. Int J Radiat Oncol Biol Phys. 2016;96:S191–S2.
Parmar C, Leijenaar RT, Grossmann P, Velazquez ER, Bussink J, Rietveld D, et al. Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Sci Rep. 2015;5:11044.
Parmar C, Grossmann P, Rietveld D, Rietbergen MM, Lambin P, Aerts HJ. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front Oncol. 2015;5:272.
Sala E, Mema E, Himoto Y, Veeraraghavan H, Brenton J, Snyder A, et al. Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol. 2017;72:3–10.
Radiology ESo. Medical imaging in personalised medicine: a white paper of the research committee of the European Society of Radiology (ESR). Insights Imaging. 2011;2:621–30.
Gutman DA, Dunn WD, Grossmann P, Cooper LA, Holder CA, Ligon KL, et al. Somatic mutations associated with MRI-derived volumetric features in glioblastoma. Neuroradiology. 2015;57:1227–37.
Banerjee S, Wang DS, Kim HJ, Sirlin CB, Chan MG, Korn RL, et al. A computed tomography radiogenomic biomarker predicts microvascular invasion and clinical outcomes in hepatocellular carcinoma. Hepatology. 2015;62:792–800.
Sadot E, Simpson AL, Do RK, Gonen M, Shia J, Allen PJ, et al. Cholangiocarcinoma: correlation between molecular profiling and imaging phenotypes. PLoS One. 2015;10:e0132953.
Yamamoto S, Han W, Kim Y, Du L, Jamshidi N, Huang D, et al. Breast cancer: radiogenomic biomarker reveals associations among dynamic contrast-enhanced MR imaging, long noncoding RNA, and metastasis. Radiology. 2015;275:384–92.
Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. J Magn Reson Imaging. 2015;42:902–7.
Li H, Zhu Y, Burnside ES, Huang E, Drukker K, Hoadley KA, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016;2:16012. https://doi.org/10.1038/npjbcancer.2016.12.
Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo W-L, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–91.
Nam H, Chung BC, Kim Y, Lee K, Lee D. Combining tissue transcriptomics and urine metabolomics for breast cancer biomarker identification. Bioinformatics. 2009;25:3151–7.
Komurov K, Tseng JT, Muller M, Seviour EG, Moss TJ, Yang L, et al. The glucose-deprivation network counteracts lapatinib-induced toxicity in resistant ErbB2-positive breast cancer cells. Mol Syst Biol. 2012;8:596.
Jayaram S, Gupta MK, Raju R, Gautam P, Sirdeshmukh R. Multi-omics data integration and mapping of altered kinases to pathways reveal gonadotropin hormone signaling in glioblastoma. OMICS. 2016;20:736–46.
Dazert E, Colombi M, Boldanova T, Moes S, Adametz D, Quagliata L, et al. Quantitative proteomics and phosphoproteomics on serial tumor biopsies from a sorafenib-treated HCC patient. Proc Natl Acad Sci. 2016;113:1381–6.
Chan KA, Jiang P, Chan CW, Sun K, Wong J, Hui EP, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci. 2013;110:18761–8.
Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra54-ra54.
Chan KA, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211–24.
Munoz J, Kurzrock R. Targeted therapy in rare cancers—adopting the orphans. Nat Rev Clin Oncol. 2012;9:631–42.
Kou T, Kanai M, Matsumoto S, Okuno Y, Muto M. The possibility of clinical sequencing in the management of cancer. Jpn J Clin Oncol. 2016;46:399–406.
Lee J-M, Han JJ, Altwerger G, Kohn EC. Proteomics and biomarkers in clinical trials for drug development. J Proteome. 2011;74:2632–41.
Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;2001:1031–7.
Demetri GD, Von Mehren M, Blanke CD, Van den Abbeele AD, Eisenberg B, Roberts PJ, et al. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–80.
Greenhalgh J, Dwan K, Boland A, Bates V, Vecchio F, Dundar Y, et al. First-line treatment of advanced epidermal growth factor receptor (EGFR) mutation positive non-squamous non-small cell lung cancer. Cochrane Libr. 2016. https://doi.org/10.1002/14651858.CD010383.
Blumenschein GR, Saintigny P, Liu S, Kim ES, Tsao AS, Herbst RS, et al. Comprehensive biomarker analysis and final efficacy results of sorafenib in the BATTLE trial. Clin Cancer Res. 2013;19:6967–75.
Song P, Gao J, Inagaki Y, Kokudo N, Hasegawa K, Sugawara Y, et al. Biomarkers: evaluation of screening for and early diagnosis of hepatocellular carcinoma in Japan and China. Liver Cancer. 2013;2:31–9.
Song P, Feng X, Zhang K, Song T, Ma K, Kokudo N, et al. Perspectives on using des-γ-carboxyprothrombin (DCP) as a serum biomarker: facilitating early detection of hepatocellular carcinoma in China. Hepatobiliary Surg Nutr. 2013;2:227.
Acknowledgements
The authors acknowledge financial support from the National Natural Science Foundation of China (Grant No. 81572278 and 81272798 to X.Z.), the Hunan Provincial Natural Science Foundation of China (Grant No. 14JJ7008 to X. Z.), and the Xiangya Hospital Funds for Talent Introduction (to X.Z.), and China “863” Plan Project (Grant No. 2014AA020610-1 to X. Z.).
Author information
Authors and Affiliations
Contributions
M.L. collected and analyzed references, and wrote manuscript draft. X.Z. conceived the concept, collected references, designed and critically revised the manuscript, and trained M.L. regarding omics, systems biology, personalized medicine, and precision medicine. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this article.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lu, M., Zhan, X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA Journal 9, 77–102 (2018). https://doi.org/10.1007/s13167-018-0128-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13167-018-0128-8