Abstract
Strong evidence implicates maternal environmental exposures in contributing to adverse outcomes during pregnancy and later in life through the developmental origins of health and disease hypothesis. Recent research suggests these effects are mediated through the improper regulation of DNA methylation in offspring tissues, specifically placental tissue, which plays a critical role in fetal development. This article reviews the relevant literature relating DNA methylation in multiple tissues at or near delivery to several prenatal environmental toxicants and stressors, including cigarette smoke, endocrine disruptors, heavy metals, as well as maternal diet. These human studies expand upon previously reported outcomes in animal model interventions and include effects on both imprinted and non-imprinted genes. We have also noted some of the strengths and limitations in the approaches used, and consider the appropriate interpretation of these findings in terms of their effect size and their relationship to differential gene expression and potential health outcomes. The studies suggest an important role of DNA methylation in mediating the effects of the intrauterine environment on children’s health and a need for additional research to better clarify the role of this epigenetic mechanism as well as others.
Similar content being viewed by others
Introduction
The developmental origins of health and disease hypothesis postulates that environmental factors encountered in utero may heavily influence disease susceptibility and outcomes in adult offspring [1]. Understanding developmental plasticity in response to environmental exposures and the potential epigenetic mechanisms of these influences holds great promise for both prediction of disease risk and novel disease intervention. The most widely studied and best understood mechanism of action for epigenetic influence on health and disease is DNA methylation. Mammalian DNA methylation occurs predominantly at cytosine residues immediately followed by a guanine, which are known as CpG sites. CpG site methylation occurs through either de novo or maintenance methylation actively performed by DNA methyltransferase (DNMT) proteins (reviewed in [2]). DNA methylation levels may change throughout an individual’s life; however, a large portion of the epigenome is established during fetal development [3].
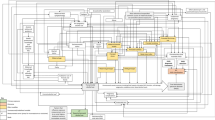
Studies investigating alterations in DNA methylation as a result of prenatal or neonatal exposures to environmental stimuli have begun to shed light on how different factors affect gene expression in different tissues including fetal cord blood and placental tissue. Within studies pertaining to DNA methylation, both highlighted here as well as outside of the scope of this review, it is important to keep in mind the highly cell-specific nature of DNA methylation that has been described by our lab, as well as others [4, 5, 6••]. This cellular specificity has important implications for studies involving DNA methylation. First, studies of environmental impact on epigenetics in non-pathologic tissues (such as the placenta or infant cord blood) often observe relatively small effect sizes, reflected in differences in methylation of only a few percent. This is in sharp contrast to the differences in DNA methylation observed within pathologic tissues such as cancerous tumors, where methylation in promoter regions may differ by orders of magnitude between the tumor and its relevant non-diseased counterpoint. Yet, it is critical to recognize that tumors are clonal tissue samples, and likely the DNA methylation measured within those samples may represent the status in a large majority of the cells within the tumor. In non-pathologic samples, variation in methylation may occur in only some small percentage of cells, and so when a subset of the tissue is measured, the result is a diluted measurable difference (Fig. 1). This would not indicate a lack of merit to these findings, but that the cellular specificity is important.
Analysis of healthy non-pathologic tissue samples frequently yields very small differences between treatment and reference population in comparison with analysis of tumor versus healthy tissue. a Much of this is due to the propagation of cells with aberrant methylation via clonal expansion in cancerous tissues, yielding a higher proportion of cells within the tissue having the altered methylation state. b Following sampling of the tissues, downstream analysis of the methylation pattern of the tissues of interest may yield a map of the methylome. c Though the non-pathologic tissues may in fact hold a subset of cells with varied methylation in comparison to a reference population, these differences will appear to be much smaller than an analysis involving cancerous tissues, again, due to the differential proportion of cells with varied methylation
Along those lines, researchers must be cognizant of the mixed nature of non-pathologic samples including peripheral blood, the placenta, or any number of other tissues. As DNA methylation defines cellular differentiation, examination of human tissue samples that include multiple cell types need to correct for confounding of the results due to the multiple types of cells present within a sample. Methods to perform this correction based upon the use of a known referent population of cells [7] or utilizing a reference-free correction [8••] have been developed to address these issues. Finally, due to methylation’s tissue specificity, the role or function of any tissue being considered for DNA methylation must be taken into account both when selecting what cell type to investigate as well as in the interpretation of the subsequent data collected. This is often impractical in a population context, where target organs and tissue may not be readily accessible, but even accessible tissues can be utilized as long as the appropriate understanding of the potential relevance of any findings is considered.
With this in mind, the placenta may be considered an ideal candidate for investigation as it is a mediator of nutrient and waste exchange, regulates interaction with the maternal immune system, acts as a neuroendocrine organ producing hormones and growth factors necessary for fetal development, and regulates fetal exposure to intrinsic and exogenous exposures present in the maternal circulation, making it a master regulator for the fetal environment, and thus a focus of this review [9–11]. These characteristics make the placenta a promising target for investigation into the interplay between environment and fetal development, as it may serve not only as a biomarker but also underlies mechanisms responsible for fetal programming.
Animal toxicological models have focused researchers’ attention on specific environmental factors that will be discussed more extensively throughout this review, including fetal exposure to cigarette smoke [12, 13]; endocrine disruptors, such as bisphenol A [14–16]; heavy metals [17, 18]; and maternal nutrition including obesity and diabetes [19]. We aim to address the role of these different environmental exposures (Fig. 2) on the human fetus or neonate and the subsequent alteration of DNA methylation patterns in varying tissue samples (summarized in Table 1). Special attention will be paid to the role of intrauterine exposures on placental methylation and the potential link between environment and gene expression. In addition, we highlight the effect of environmental stimuli on imprinting, as the important role that imprinting control regions may play in development has been reviewed extensively [41, 42]. Finally, we aim to address the knowledge gaps underlying human epidemiological studies focusing on the intrauterine environment as well as our understanding of tissue-specific alterations in methylation in response to these environmental stimuli.
Cigarette Smoke
Prenatal exposure to cigarette smoke has long been linked with deleterious birth phenotypes including fetal growth restriction [43], preterm birth [44], and negative outcomes for women with pre-eclampsia [45], as well as long-term health consequences including those of psychological health [46], asthma [47], and childhood obesity [48, 49]. In addition to clinical associations between smoking and infant and child health, recent studies have identified altered methylation status of genes following exposure during pregnancy to cigarette smoke, potentially linking it to altered gene expression later in life. An early implication of maternal smoking during pregnancy (MSDP) as a mediator of fetal DNA methylation came from investigations on buccal epithelium utilizing the Illumina GoldenGate Assay for Methylation for gene-specific examinations of DNA methylation. This study also included using pyrosequencing following bisulfite conversion of repetitive elements such as LINE-1 and AluYb8 (Alu) methylation on DNA obtained from buccal cells [20]. While LINE-1 showed no alterations in methylation in this children’s cohort, Alu methylation was significantly lower in smoke-exposed children. Alu segments are the most abundant repetitive element within the human genome, and transcription of these regions, due to the presence of transcription factor sequence motifs, play an important role in regulating expression of adjacent genes [50], acting as an important regulator of cellular health. In addition, eight candidate regions altered by MSDP were identified, including tyrosine-protein kinase receptor UFO (AXL) and protein tyrosine phosphatase, receptor type, O (PTPRO), both of which play an important role in cellular proliferation. Within a similar time frame, ELISA-based measurements of global methylation levels on DNA from the umbilical cord serum also linked MSDP to hypomethylation from prenatally smoke-exposed samples [21]. These initial studies were an early indication into the potential role of intrauterine smoke exposure on DNA methylation [20, 21].
A targeted, candidate gene approach utilizing bisulfite sequencing to investigate placental methylation and its relationship with gene expression following MSDP conducted by Suter et al. (2010) identified aberrant DNA methylation at CpG sites adjacent to the cytochrome P450, family 1, member A1 (CYP1A1) gene. These alterations in methylation were highly correlated with CYP1A1 gene expression, suggesting a causal link between smoking and transcription rates mediated by DNA methylation. CYP1A1 acts as an important player in metabolism of harmful carcinogenic elements found in cigarette smoke, such as polycyclic aromatic hydrocarbons [22]. In a separate follow-up study, epigenome-wide promoter methylation was investigated in placental tissue to determine its correlation with gene expression and the relationship of alterations of both to intrauterine smoke exposure. This revealed a number of CpG sites with discordant methylation status due to maternal smoke exposure displaying a modest correlation to altered gene expression [23]. Pathway analysis of the genes differentially expressed in smoke-exposed infants demonstrated enrichment for oxidative stress-associated genes. In addition, maternal smoking-associated differential methylation at as few as six CpG sites correlated with a linear reduction in birth weight.
An area of increased scrutiny in epigenetic studies is the role of DNA methylation in brain development and the subsequent role in behavioral outcomes of offspring. This can be found highlighted in studies focusing on the role of MSDP on differential neurodevelopment of offspring and the link to epigenetic variation. Exposure to maternal smoke was linked to increased infant blood methylation in the gene body of brain-derived neutrophic factor-6 (BDNF) [24]. BDNF is involved in memory formation, suggesting in utero exposure may be capable of causing epigenetic alteration to fetal brain development. Alterations to BDNF may have a role in long-term behavioral consequences such as synapse structure and regulation [51, 52], bipolar disorder [53], and stress responsiveness [54]. It is important to note however that this study utilized blood as a proxy for brain methylation, which has not been correlated, highlighting the difficulty in ascertaining the role of methylation on human brain development. In addition, BDNF methylation and expression has increasingly been scrutinized as a factor in broader behavioral outcomes including depression and several psychiatric disorders, highlighting the importance of its fetal regulation in later life outcomes [55, 56].
Maternal smoking also appears to influence the infant stress response as demonstrated by a lower level of cortisol production under both basal and stress conditions in exposed offspring as compared to unexposed. Examination of the placental glucocorticoid receptor (NR3C1) promoter region by bisulfite sequencing revealed altered methylation levels associated with maternal smoking in line with differential saliva cortisol levels [26]. Other studies utilizing Illumina array technology have investigated the role of maternal smoking on methylation. Maternal smoking has been associated with aberrant placental methylation of the runt-related transcription factor 3 (RUNX3) gene [25], as well as cord blood aryl hydrocarbon receptor repressor (AHRR) and CYP1A1 [57•].
One factor leading to increased difficulty in analyzing epigenetic studies is the tissue-specific nature of methylation and the potentially different responses in varying neonatal tissues that may confound results between different studies. This was nicely investigated in a study conducted by Novakovic et al. (2013) in which the authors aimed to confirm previously reported [57•, 58, 59] alterations in methylation of the AHRR gene in multiple fetal tissues [6••]. To accomplish this, methylation of the AHRR gene was quantified in cord blood mononuclear cells, buccal epithelium, as well as placental tissue, and monitored for stability in the individuals at 18 months of age. The authors first reported variable methylation levels across the tissue types, specifically in the CpG island region of the AHRR gene from placental tissue. In addition, the data confirmed that smoking throughout pregnancy altered the methylation status of the AHRR gene in neonatal blood. Methylation levels were comparable between neonatal and 18-month sampling times, suggesting stability of methylation with the potential for long-term consequences.
Taking the sum of the data available on maternal smoking, it becomes clear that intrauterine exposure to cigarette smoke plays a major role in infant health outcomes. A strong case may be made that alterations in DNA methylation are a mechanistic control point for these adverse health outcomes, including neurodevelopmental issues.
Endocrine Disruptors
Though the definition of endocrine disrupting chemicals (EDCs) is broad, and at times controversial [60], they are generally considered to be a wide-ranging group of compounds that, at certain doses, act to alter the function of hormones within mammals. Chemicals that fall within this category include phenols such as bisphenol A (BPA), or phthalates, both of which are important plasticizers. These compounds frequently enter the environment and food chain via manufacturing waste, pesticide use, or water contamination. While endocrine disruptors such as BPA are fairly ubiquitous in everyday life, their role in altering human health has been somewhat contentious [61], with studies on the role of prenatal exposure in humans only recently becoming more prominent. BPA has conclusively been shown to affect the epigenome however through the alteration of the metastable Agouti allele in mice. Genetically identical individuals differ in coat color based upon DNA hypomethylation at the Agouti gene as a result of BPA consumption. Further linking this alteration to the actions of DNA methylation, this phenomenon may be reversed by nutritional supplementation with methyl donors (folic acid) [62]. Interestingly, animal models have demonstrated that the liver undergoes a 40 % reduction in the ability to metabolize BPA into its inactivated state during pregnancy [63], suggesting an acute susceptibility to endocrine disruptors during this period for both the mother and fetus. This is supported by human studies that found that pregnant women excreted 26 % greater levels of BPA in their urine as compared to pre-pregnancy [64]. These findings would suggest that exposure to endocrine disruptors during pregnancy have a great potential for harm, of which epigenetic mechanisms may be playing an important role.
Though limited in scope, several recent publications have implicated prenatal BPA exposure as a mediator of DNA methylation in human populations. Nahar et al. (2014) demonstrated that in utero exposure to bisphenol A altered expression of xenobiotic metabolizing enzymes (XMEs) in human fetal liver tissue. Of the approximately 160 XMEs investigated, 14 displayed reduced expression as a result of increased prenatal BPA exposure. To further elucidate the mechanism linking BPA exposure and expression of these XMEs, promoter methylation levels were measured for carboxylesterase 2 (CES2), catechol-O-methyltransferase (COMT), and sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 (SULT2A1). This yielded both site-specific (COMT) and average methylation (SULT2A1) differences in response to BPA, implying causality between exposure and XME expression [27].
In addition to BPA, phthalate and phenol exposure has recently been investigated for their role in altering DNA methylation following prenatal exposure in the first trimester. Exposure to EDCs was quantified by proxy measurement of maternal urine levels, and fetal DNA methylation was measured in placental tissue. Increasing maternal levels of the sum of phthalate metabolites measured were negatively correlated with placental methylation of the imprinted genes H19 and several regions of insulin-like growth factor 2 (IGF2) [28••]. H19 and IGF2 form a gene cluster on human chromosome 11, and have been shown to be heavily regulated by epigenetic mechanisms (common among imprinted genes) with IGF2 acting as an important fetal growth factor [65]. Though endocrine disruptor exposure appeared to alter methylation of these genes, the changes could not be directly linked to variation in their allele-specific expression or any birth outcomes. These findings suggest that EDC exposure may alter placental methylation, though a more thorough investigation is required to determine its role in infant health and birth outcomes.
Heavy Metal Exposure
Heavy metals may be broadly defined as chemicals with a density much greater than that of water, and although some heavy metals including iron, copper, and zinc are essential to a balanced diet, many heavy metals pose potential health hazards, including arsenic, cadmium, mercury, and lead. As maternal metal exposure has received more scrutiny, evidence has emerged to suggest that heavy metals may induce epigenetic changes in fetal tissues.
Arsenic
Arsenic exposure for pregnant women may originate from the use of unregulated wells for drinking water as well as rice consumption, with levels of exposure varying widely depending on the population and region studied [66]. Arsenic consumption at moderate to high levels has been associated with increased infant mortality and low-birth weight [67, 68]. Several hypotheses exist as to the mechanisms through which arsenic exposure would mediate DNA methylation levels. These include competition for DNA methyltransferase binding or depletion of methyl donors [69, 70], leading to general transcriptional deregulation. Investigations on the relationship between maternal arsenic exposure and global DNA methylation in both umbilical cord blood and maternal leukocytes from a Bangladeshi population with a range of exposures beyond what would be seen in developed countries suggested that arsenic exposure plays a role in altering methylation levels for both mother and child. Measurements of LINE-1 methylation levels showed increases of 1.36 and 1.08 % in umbilical cord blood and maternal leukocytes, respectively; however, there were no differences seen in Alu or p53 methylation levels [29]. In a more recent study in New Hampshire, where 10–15 % of the population relies on water contaminated with arsenic beyond the EPA standard, genome-wide methylation was examined by using the Illumina HumanMethylation450 BeadChip array in umbilical cord blood and corrected the data based upon leukocyte subtype proportions. Based on this genome-wide DNA methylation data, this study identified an increase in the proportion of CD8+ T cells as well as differential methylation in regions across the genome. There was an enrichment for arsenic-associated DNA methylation in CpG islands, suggesting that this variation may be functional [31•]. The finding of reduced CD8+ T cells was later replicated, again using cord blood, in a more highly exposed Bangladeshi population [30•].
Cadmium
Cadmium exposure primarily occurs through cigarette smoke and dietary intake, with high levels found in offal meats, crustaceans, mollusks, and some leafy greens [71, 72]. Cadmium is a partial transplacental toxic metal, accumulating in the placenta, and has been implicated as a potential cause of adverse birth outcomes [73]. Specifically, in a rat model, prenatal exposure to cadmium altered methylation of the hepatic glucocorticoid receptor [17], which may correlate with human data demonstrating an altered glucocorticoid response in human placental trophoblasts following cadmium exposure [74]. A lack of epigenetic data on the role of cadmium in glucocorticoid-related stress responses in utero represents a major gap in current understanding of the molecular mechanisms of intrauterine cadmium exposure.
Epigenetic data has also implicated maternal cadmium exposure as a negative driver of fetal birth weight. A prospective cohort study performed by Kippler et al. (2012) demonstrated a negative linear relationship between maternal urinary cadmium levels with birth weight and head circumference in girls, though no association was seen for boys [73]. A follow-up study investigated the role of cadmium on cord blood DNA methylation levels using the Infinium HumanMethylation450 BeadChip. This study found that cadmium exposure could be associated with altered blood methylation; however, as was seen in the previous work, the alterations were highly sex-specific. Correlations between exposure levels and methylation in boys were predominantly positive (96 % of the top 500 CpG sites), while in girls only 29 % of the top 500 correlated CpG sites showed a positive relationship. In addition, the genes showing strong correlations to cadmium levels differed between the sexes as girls were predominantly affected in genes related to organ development, while changes in boys were in cell-death-related genes. Though the authors had previously described an inverse relationship between cadmium and birth weight in girls, this study was unable to identify specific CpG sites affecting birth weight [33]. These data would suggest that cadmium exposure is playing a critical role in DNA methylation levels, potentially affecting gene expression in both boys and girls, though in different genomic regions, and may be a factor in birth weight outcomes.
Lead
Though lead has been shown to be detrimental to the health of a developing fetus [75–77], very little research has been conducted in determining the role of maternal exposure on DNA methylation. An in vitro study utilizing isolated embryonic stem cells (ESCs) exposed to a biologically relevant dose of lead monitored the effects on DNA methylation and neuronal differentiation potential. Lead exposure altered methylation in multiple neuronal-related genes, and neural progenitor cells from these ESCs differentiated into neurons with aberrant morphology [34]. This result may be due to altered methyltransferase expression following prenatal lead exposure, as described in a rodent model examining the effects of lead on neural development [78]. Though a number of publications have detailed the detrimental outcomes associated with prenatal lead exposure, there is a void in the literature examining the molecular basis underlying these pathologies, specifically how DNA methylation may be involved. This is likely due to the difficultly associated with obtaining relevant tissue samples to neural development, again highlighting the importance of representative proxy tissues, such as the placenta.
Maternal Diet
Though we have seen that what a fetus is exposed to via maternal diet is likely playing a role in altering DNA methylation, there is evidence that how much is consumed during pregnancy may also be important in development. Though initial investigation of diet identified the role of famine on fetal outcome and DNA methylation [79, 80], a more relevant branch of nutritional research to the modern US population [81] has begun to focus on maternal obesity and gestational diabetes mellitus (GDM). A study conducted by Ruchat et al. (2013) found that GDM during pregnancy altered DNA methylation in both offspring placental tissue and cord blood. These changes, measured by Infinium HumanMethylation450 BeadChip, moderately overlapped (1029 of approximately 3500 differentially methylated genes) between the two tissues examined. Notably, the genes displaying differential methylation levels between GDM-exposed children and controls were enriched for genes involved in metabolic disease pathways, suggesting that maternal diabetes epigenetically alters offspring metabolism [35•].
This role of maternal metabolic condition on offspring epigenetics is highlighted by several studies that have focused on the role of maternal obesity on methylation within the metabolically important leptin (LEP) gene. An investigation into the tissue-specific methylation of leptin examined both maternal and infant cord blood as well as the placenta. The study found a correlation between maternal obesity and cord blood leptin methylation levels, as well as lower levels of methylation in infants born to pre-pregnancy obese mothers. In both the placenta and cord blood, infants born small for gestational age had significantly higher LEP methylation [82•]. In a similar study focusing specifically on the role of GDM within the placenta, maternal pre-pregnancy obesity and GDM status led to significantly higher LEP methylation levels [36].
Investigations have also implicated gestational diabetes mellitus on altered imprinted gene methylation levels. Placental tissue and cord blood were analyzed for GDM-exposed babies and non-exposed controls across seven imprinted genes as well as in LINE-1 and Alu as genome-wide methylation proxies. Of the genes analyzed, the maternally imprinted gene MEST displayed markedly reduced methylation in GDM-exposed babies compared to controls. This was also seen in a separate group of morbidly obese adults, analyzed in the same data set, suggesting a correlated relationship and mechanistic link between the ability of an excessive diet to alter methylation in both a developing fetus as well as a mature adult [38]. Though these studies focused on specific genes or pathways, analysis on a more global scale across several studies has demonstrated the potentially broad impact of GDM on epigenome-wide methylation [83, 84]. Taken together, this data strongly implicates gestational diabetes mellitus as modulators of offspring metabolic pathways, potentially predisposing them to metabolic diseases.
Dietary folate acts as single carbon methyl donor, and restricted intake of this carbon source may lead to negative birth outcomes [85]. The correlation between nutritional sources of methyl donors and infant phenotype points towards a pivotal role of folate in DNA methylation status. For example, maternal periconceptual nutritional intake in The Gambia, with folate as a model cofactor, had a strong effect on offspring lymphocyte DNA methylation with a correlation to levels of several plasma biomarkers, including methionine, choline, and S-adenosylmethionine, among others [86]. A closer examination of folate supplementation after 12 weeks of pregnancy investigated methylation status of several imprinted genes in cord blood. Supplementation was associated with higher IGF2 methylation and a reduction in both PEG3 and LINE-1 methylation status [39]. Aberrant methylation at imprinted sites has been reported elsewhere, with several studies linking the alteration with subsequent infant birth weight [40, 87]. Interestingly, the mechanism of altered DNA methylation in the presence or absence of excess folate at imprinting sites may not be due to methyl group bioavailability, but to altered regulation of zinc finger protein-57 (ZFP57). A recent investigation utilizing an epigenome-wide approach looked at methylation status of neonate CD4+ T cells and antigen-presenting cells exposed to either a high or low maternal folate diet. Among the regions differentially methylated, the largest effect was seen in a 923 bp window 3 kb upstream of ZFP57, in which a high-folate diet correlated with hypomethylation [88]. ZFP57 plays a critical role in the establishment and maintenance of imprinting-associated DNA methylation, establishing a link between maternal folate consumption and genetic imprinting [89, 90]. The critical link between imprinting and placental biology has previously been reviewed, with a focus on pathology following alterations in normal imprinting patterns [91].
The World Health Organization (WHO), among others, has issued a recommendation of at least 0.4 mg of folate intake periconceptually in order to potentially prevent the development of neural tube defects [92]. A better understanding of whether this recommendation is adequate (or excessive [93, 94]) and how dietary folate may best be utilized to reduce adverse birth outcome would greatly benefit clinical obstetricians.
Conclusions
This review has covered the role of prenatal and neonatal environmental exposures with alterations in DNA methylation. Among the studies mentioned in this review, placental tissue has emerged as a popular candidate for analysis of DNA methylation. Appropriate placental gene expression is paramount to fetal regulation during pregnancy, and alterations from this have been linked to pre-eclampsia, intrauterine growth restriction, gestational diabetes mellitus, and trophoblastic disease [95–99]. The strong association between gene expression and DNA methylation point towards a potentially significant mechanism of prenatal programming.
While an understanding of the mechanisms controlling DNA methylation and gene expression in the placenta would be a major step towards the understanding of adverse pregnancy outcomes, a direct link to environmental exposures predisposing pregnancies to complications would aid in the reduction or elimination of these issues. With all of this taken into account, there is currently a void of knowledge in the linkages between environmental exposures and disease. More work needs to be done utilizing large birth cohorts with expansive clinical data on maternal exposures along with birth outcomes and infant methylation status. Ideally, these data sets would include plans for longitudinal follow-ups of infant health. The continuing improvement of our understanding of the intrauterine environment, methylation, and disease will continue to progress our utilization of risk predictors and biomarkers to help clinicians better diagnose and treat complex disease.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Hanson MA, Gluckman PD. Developmental origins of health and disease: new insights. Basic Clin Pharmacol Toxicol. 2008;102(2):90–3.
Jeltsch A, Jurkowska RZ. New concepts in DNA methylation. Trends Biochem Sci. 2014;39(7):310–8.
Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–32.
Armstrong DA et al. Global and gene-specific DNA methylation across multiple tissues in early infancy: implications for children’s health research. FASEB J. 2014;28(5):2088–97.
Christensen B et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5(8):e1000602.
Novakovic B et al. Postnatal stability, tissue, and time specific effects of AHRR methylation change in response to maternal smoking in pregnancy. Epigenetics. 2013;9(3):377–86. Demonstrates the tissue specific nature of the alterations associated with cigarette smoke as well as the longevity of these changes.
Houseman E et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13(1):86.
Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30(10):1431–9. Introduces a technique that is able to account for cellular proportions when utilizing the 450K methylationarray technology. This allows for increased power in detection of true positives due to the ability to correct for different cell proportions across samples.
Novakovic B, Saffery R. The ever growing complexity of placental epigenetics—role in adverse pregnancy outcomes and fetal programming. Placenta. 2012;33(12):959–70.
Maccani MA, Marsit CJ. Epigenetics in the placenta. Am J Reprod Immunol. 2009;62(2):78–89.
Banister CE et al. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. Epigenetics. 2011;6(7):920–7.
Rehan V et al. Perinatal nicotine exposure induces asthma in second generation offspring. BMC Medicine. 2012;10(1):129.
Lawrence J et al. Foetal nicotine exposure causes PKCε gene repression by promoter methylation in rat hearts. Cardiovasc Res. 2011;89(1):89–97.
Dhimolea E et al. Prenatal exposure to BPA alters the epigenome of the rat mammary gland and increases the propensity to neoplastic development. PLoS One. 2014;9(7):e99800.
Jefferson W et al. Persistently altered epigenetic marks in the mouse uterus after neonatal estrogen exposure. Mol Endocrinol. 2013;27(10):1666–77.
Kundakovic M et al. Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proc Natl Acad Sci. 2013;110(24):9956–61.
Castillo P et al. Impact of cadmium exposure during pregnancy on hepatic glucocorticoid receptor methylation and expression in rat fetus. PLoS One. 2012;7(9):e44139.
Tsang V et al. The epigenetic effects of a high prenatal folate intake in male mouse fetuses exposed in utero to arsenic. Toxicol Appl Pharmacol. 2012;264(3):439–50.
Ge Z-J et al. Maternal obesity and diabetes may cause DNA methylation alteration in the spermatozoa of offspring in mice. Reprod Biol Endocrinol. 2014;12(1):29.
Breton CV et al. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180(5):462–7.
Guerrero-Preston R et al. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics. 2010;5(6):539–46.
Suter M et al. In utero tobacco exposure epigenetically modifies placental CYP1A1 expression. Metabolism. 2010;59(10):1481–90.
Suter M et al. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics. 2011;6(11):1284–94.
Toledo-Rodriguez M et al. Maternal smoking during pregnancy is associated with epigenetic modifications of the brain-derived neurotrophic factor-6 exon in adolescent offspring. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(7):1350–4.
Maccani JZ et al. Placental DNA methylation alterations associated with maternal tobacco smoking at the RUNX3 gene are also associated with gestational age. Epigenomics. 2013;5(6):619–30.
Stroud LR et al. Maternal smoking during pregnancy and infant stress response: test of a prenatal programming hypothesis. Psychoneuroendocrinology. 2014;48:29–40.
Nahar MS et al. Bisphenol A-associated alterations in the expression and epigenetic regulation of genes encoding xenobiotic metabolizing enzymes in human fetal liver. Environ Mol Mutagen. 2014;55(3):184–95.
LaRocca J et al. The impact of first trimester phthalate and phenol exposure on IGF2/H19 genomic imprinting and birth outcomes. Environ Res. 2014;133:396–406. Demonstrates the association between early pregnancy phthalate exposure and alterations to DNAmethylation at the IGF2/H19 gene cluster. This is an imprinted region and critical for proper development.
Kile ML et al. Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ Health Perspect. 2012;120(7):1061–6.
Kile ML et al. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics. 2014;9(5):774–82. Highlights the role of arsenic exposure in utero to not only DNA methylation, but also to leukocyte proportions.
Koestler D et al. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ Health Perspect. 2013;121(8):971–7. Associates in utero arsenic exposure with broad changes to cord blood DNA methylation levels.
Broberg K et al. Arsenic exposure in early pregnancy alters genome-wide DNA methylation in cord blood, particularly in boys. J Dev Orig Health Dis. 2014;5(4):288–98.
Kippler M et al. Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics. 2013;8(5):494–503.
Senut M-C et al. Lead exposure disrupts global DNA methylation in human embryonic stem cells and alters their neuronal differentiation. Toxicol Sci. 2014;139(1):142–61.
Ruchat S-M et al. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics. 2013;8(9):935–43. Important for its demonstration that not only does maternal gestational diabetes affect infant DNAmethylation, but that this is occurring predominantly within genes that would have a subsequent role in the infant’s nutritional health.
Lesseur C et al. Maternal obesity and gestational diabetes are associated with placental leptin DNA methylation. Am J Obstet Gynecol. 2014;211(6):654.e1-9.
Houde A-A et al. Placental lipoprotein lipase DNA methylation levels are associated with gestational diabetes mellitus and maternal and cord blood lipid profiles. J Dev Orig Health Dis. 2014;2(5):132–41.
El Hajj N et al. Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes. 2013;62(4):1320–8.
Haggarty P et al. Folate in pregnancy and imprinted gene and repeat element methylation in the offspring. Am J Clin Nutr. 2013;97(1):94–9.
Hoyo C et al. Erythrocyte folate concentrations, CpG methylation at genomically imprinted domains, and birth weight in a multiethnic newborn cohort. Epigenetics. 2014;9(8):1120–30.
Lawson HA, Cheverud JM, Wolf JB. Genomic imprinting and parent-of-origin effects on complex traits. Nat Rev Genet. 2013;14(9):609–17.
Plasschaert RN, Bartolomei MS. Genomic imprinting in development, growth, behavior and stem cells. Development. 2014;141(9):1805–13.
Varvarigou A, Fouzas S, Beratis N. Effect of prenatal tobacco smoke exposure on fetal growth potential. J Perinat Med. 2010;38(6):683–7.
Aliyu M et al. Intrauterine exposure to tobacco and risk of medically indicated and spontaneous preterm birth. Am J Perinatol. 2010;27(5):405–10.
Miller EC et al. The risk of adverse pregnancy outcomes is increased in preeclamptic women who smoke compared with nonpreeclamptic women who do not smoke. Am J Obstet Gynecol. 2010;203(4):334.e1–8.
Knopik VS et al. The epigenetics of maternal cigarette smoking during pregnancy and effects on child development. Dev Psychopathol. 2012;24(Special Issue 04):1377–90.
Burke H et al. Prenatal and passive smoke exposure and incidence of asthma and wheeze: systematic review and meta-analysis. Pediatrics. 2012;129(4):735–44.
Riedel C et al. Parental smoking and childhood obesity: higher effect estimates for maternal smoking in pregnancy compared with paternal smoking—a meta-analysis. Int J Epidemiol. 2014;43(5):1593–606.
Wang L et al. Independent and joint effects of prenatal maternal smoking and maternal exposure to second-hand smoke on the development of adolescent obesity: a longitudinal study. J Paediatr Child Health. 2014;50(11):908–15.
Häsler J, Samuelsson T, Strub K. Useful ‘junk’: Alu RNAs in the human transcriptome. Cell Mol Life Sci. 2007;64(14):1793–800.
Zagrebelsky M, Korte M. Form follows function: BDNF and its involvement in sculpting the function and structure of synapses. Neuropharmacology. 2014;76(Part C(0)):628–38.
Ninan I. Synaptic regulation of affective behaviors; role of BDNF. Neuropharmacology. 2014;76(Part C(0)):684–95.
Frey BN et al. Biomarkers in bipolar disorder: a positional paper from the International Society for Bipolar Disorders Biomarkers Task Force. Aust N Z J Psychiatry. 2013;47(4):321–32.
Mahan AL, Ressler KJ. Fear conditioning, synaptic plasticity and the amygdala: implications for posttraumatic stress disorder. Trends Neurosci. 2012;35(1):24–35.
Ikegame T et al. DNA methylation of the BDNF gene and its relevance to psychiatric disorders. J Hum Genet. 2013;58(7):434–8.
Song Y et al. Altered DNA methylation status of human brain derived neurotrophis factor gene could be useful as biomarker of depression. Am J Med Genet B Neuropsychiatr Genet. 2014;165(4):357–64.
Joubert BR et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect. 2012;120(10):1425–31. A study that broadly implicates maternal smoking with DNA methylation utilizing the genome-wide 450K methylation array.
Philibert RA, Beach SRH, Brody GH. Demethylation of the aryl hydrocarbon receptor repressor as a biomarker for nascent smokers. Epigenetics. 2012;7(11):1331–8.
Philibert R et al. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clinical Epigenetics. 2013;5(1):19.
Foster WG, Agzarian J. Toward less confusing terminology in endocrine disruptor research. J Toxicol Environ Health Part B. 2008;11(3–4):152–61.
Rubin BS. Bisphenol A: an endocrine disruptor with widespread exposure and multiple effects. J Steroid Biochem Mol Biol. 2011;127(1–2):27–34.
Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci. 2007;104(32):13056–61.
Inoue H et al. Bisphenol a glucuronidation and excretion in liver of pregnant and nonpregnant female rats. Drug Metab Dispos. 2005;33(1):55–9.
Mahalingaiah S et al. Temporal variability and predictors of urinary bisphenol A concentrations in men and women. Environ Health Perspect. 2008;116(2):173–8.
Nordin M et al. Epigenetic regulation of the Igf2/H19 gene cluster. Cell Prolif. 2014;47(3):189–99.
Gilbert-Diamond D et al. Rice consumption contributes to arsenic exposure in US women. Proc Natl Acad Sci. 2011;108(51):20656–60.
Rahman A et al. Arsenic exposure during pregnancy and size at birth: a prospective cohort study in Bangladesh. Am J Epidemiol. 2009;169(3):304–12.
Rahman A et al. Arsenic exposure and risk of spontaneous abortion, stillbirth, and infant mortality. Epidemiology. 2010;21(6):797–804. doi:10.1097/EDE.0b013e3181f56a0d.
Reichard JF, Schnekenburger M, Puga A. Long term low-dose arsenic exposure induces loss of DNA methylation. Biochem Biophys Res Commun. 2007;352(1):188–92.
Zhao CQ et al. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci. 1997;94(20):10907–12.
Järup L, Åkesson A. Current status of cadmium as an environmental health problem. Toxicol Appl Pharmacol. 2009;238(3):201–8.
Olsson I-M et al. Cadmium in blood and urine—impact of sex, age, dietary intake, iron status, and former smoking–association of renal effects. Environ Health Perspect. 2002;110(12):1185–90.
Kippler M et al. Maternal cadmium exposure during pregnancy and size at birth: a prospective cohort study. Environ Health Perspect. 2012;120(2):284–9.
Yang K et al. Cadmium reduces 11β-hydroxysteroid dehydrogenase type 2 activity and expression in human placental trophoblast cells. Am J Physiol Endocrinol Metab. 2006;290:E135–42.
Bellinger DC. Prenatal exposures to environmental chemicals and children’s neurodevelopment: an update. Saf Health Work. 2013;4(1):1–11.
Liu JA et al. Lead exposure at each stage of pregnancy and neurobehavioral development of neonates. NeuroToxicology. 2014;44:1–7.
Bellinger DC. Very low lead exposures and children’s neurodevelopment. Curr Opin Pediatr. 2008;20(2):172–7. doi:10.1097/MOP.0b013e3282f4f97b.
Schneider JS, Kidd SK, Anderson DW. Influence of developmental lead exposure on expression of DNA methyltransferases and methyl cytosine-binding proteins in hippocampus. Toxicol Lett. 2013;217(1):75–81.
Heijmans BT et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci. 2008;105(44):17046–9.
Tobi EW et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18(21):4046–53.
Mission JF, Marshall NE, Caughey AB. Obesity in pregnancy: a big problem and getting bigger. Obstet Gynecol Surv. 2013;68(5):389–99.
Lesseur C et al. Tissue-specific leptin promoter DNA methylation is associated with maternal and infant perinatal factors. Mol Cell Endocrinol. 2013;381(1–2):160–7. Demonstrates the downstream link between infant DNA methylation at the leptin gene and subsequent perinatal birth outcomes such as infant weight.
del Rosario MC et al. Potential epigenetic dysregulation of genes associated with MODY and type 2 diabetes in humans exposed to a diabetic intrauterine environment: an analysis of genome-wide DNA methylation. Metabolism. 2014;63(5):654–60.
Nomura Y et al. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod Sci. 2014;21(1):131–7.
Zeisel SH. Importance of methyl donors during reproduction. Am J Clin Nutr. 2009;89(2):673S–7S.
Dominguez-Salas P et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun. 2014;5:3746.
Vidal AC et al. Associations between antibiotic exposure during pregnancy, birth weight and aberrant methylation at imprinted genes among offspring. Int J Obes. 2013;37(7):907–13.
Amarasekera M et al. Genome-wide DNA methylation profiling identifies a folate-sensitive region of differential methylation upstream of ZFP57-imprinting regulator in humans. FASEB J. 2014;28(9):4068–76.
Zuo X et al. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J Biol Chem. 2012;287(3):2107–18.
Quenneville S et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44(3):361–72.
Tycko B. Imprinted genes in placental growth and obstetric disorders. Cytogenet Genome Res. 2006;113(1–4):271–8.
Prevention of neural tube defects. Standards for maternal and neonatal care. Geneva: World Health Organization (WHO); 2006.
Mikael LG et al. Moderately high intake of folic acid has a negative impact on mouse embryonic development. Birth Defects Res Part A: Clin Mol Teratol. 2013;97(1):47–52.
Pickell L et al. High intake of folic acid disrupts embryonic development in mice. Birth Defects Res Part A: Clin Mol Teratol. 2011;91(1):8–19.
Iglesias-Platas I et al. Altered expression of the imprinted transcription factor PLAGL1 deregulates a network of genes in the human IUGR placenta. Hum Mol Genet. 2014;23(23):6275–85.
Mandò C et al. Placental mitochondrial content and function in intrauterine growth restriction and preeclampsia. Am J Physiol Endocrinol Metab. 2014;306:E404–13.
Zhao M et al. Placental expression of VEGF is increased in pregnancies with hydatidiform mole: possible association with developing very early onset preeclampsia. Early Hum Dev. 2013;89(8):583–8.
Yang M. et al. IgG expression in trophoblasts derived from placenta and gestational trophoblastic disease and its role in regulating invasion. Immunol Res. 2014;60(1):91–104.
Nitert M et al. Increased placental expression of fibroblast growth factor 21 in gestational diabetes mellitus. J Clin Endocrinol Metab. 2014;99(4):E591–8.
Acknowledgments
This work is supported by NIH-NIMH R01MH094609, NIH-NIEHS R01ES022223, and NIH-NIEHS P01 ES022832 and by the US EPA grant RD83544201. Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the US EPA. Further, the US EPA does not endorse the purchase of any commercial products or services mentioned in the publication.
Compliance with Ethics Guidelines
ᅟ
Conflict of Interest
Benjamin B. Green and Carmen J. Marsit declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any new studies with human or animal subjects performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Environmental Epigenetics
Rights and permissions
About this article
Cite this article
Green, B.B., Marsit, C.J. Select Prenatal Environmental Exposures and Subsequent Alterations of Gene-Specific and Repetitive Element DNA Methylation in Fetal Tissues. Curr Envir Health Rpt 2, 126–136 (2015). https://doi.org/10.1007/s40572-015-0045-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40572-015-0045-0