Abstract
piRNAs, a class of small non-coding RNAs associated with PIWI proteins, have broad functions in germline development, transposon silencing, and epigenetic regulation. In diverse organisms, a subset of piRNAs derived from repeat sequences are produced via the interplay between two PIWI proteins. This mechanism, termed “ping-pong” cycle, operates among the PIWI proteins of the primordial mouse testis; however, its involvement in postnatal testes remains elusive. Here we show that adult testicular piRNAs are produced independent of the ping-pong mechanism. We identified and characterized large populations of piRNAs in the adult and postnatal developing testes associated with MILI and MIWI, the only PIWI proteins detectable in these testes. No interaction between MILI and MIWI or sequence feature for the ping-pong mechanism among their piRNAs was detected in the adult testis. The majority of MILI- and MIWI-associated piRNAs originate from the same DNA strands within the same loci. Both populations of piRNAs are biased for 5′ Uracil but not for Adenine on the 10th nucleotide position, and display no complementarity. Furthermore, in Miwi mutants, MILI-associated piRNAs are not downregulated, but instead upregulated. These results indicate that the adult testicular piRNAs are predominantly, if not exclusively, produced by a primary processing mechanism instead of the ping-pong mechanism. In this primary pathway, biogenesis of MILI- and MIWI-associated piRNAs may compete for the same precursors; the types of piRNAs produced tend to be non-selectively dictated by the available precursors in the cell; and precursors with introns tend to be spliced before processed into piRNAs.
Similar content being viewed by others
Introduction
PIWI-interacting RNAs (piRNAs) are a complex class of small non-coding RNAs of mostly 24-31 nucleotides in length that are associated with PIWI proteins. piRNA have been implicated in broad functions in germline development, transposon silencing, and epigenetic regulation1. The ping-pong mechanism has been proposed for the amplification of primary, pre-existing piRNAs, and production of secondary, new piRNAs of transposon origin in the fly, fish and mouse2,3,4,5,6,7. According to this model, a primary piRNA maternally loaded or produced by a yet-to-be identified mechanism guides its PIWI protein partner to a transcript with a complementary sequence, which leads to the production of a secondary piRNA from the targeted transcript via the “slicer” activity of the PIWI protein. The resulting secondary piRNA is then loaded onto another PIWI protein. This mechanism leads to the production of new primary piRNAs from their precursor transcripts, which eventually amplifies the pool of both primary and secondary piRNAs. This positive feedback loop is a secondary biogenesis mechanism that requires complementary transcripts to a pre-existing pool of piRNAs.
Two PIWI proteins in the mouse, MILI and MIWI2, have been proposed to engage in ping-pong for piRNA biogenesis in the primordial germ cells of 16.5 dpc testes5, where MIWI2 piRNAs are not produced in the absence of MILI. However, MIWI2 expression is not detected beyond 4 dpp8, yet the major boost of piRNA production occurs after 14 dpp9,10,11,12,13. This period corresponds to the onset of MIWI expression14, overlapping with MILI expression5,15,16 and thus raises the question of whether MIWI and MILI are engaged in the ping-pong-mediated biogenesis to produce their piRNAs. This paper focuses on addressing this question.
Results
Postnatal testicular piRNAs are largely derived from non-transposon sequences
To gain an insight into the biogenesis of piRNAs in postnatal testes, we first cloned MILI- and MIWI-associated piRNAs from 7-8 dpp, 13 dpp, and 2-month-old adult testes by immunoprecipitation of MILI and MIWI (Figure 1A), and identified them by high throughput sequencing. These time points correspond to critical stages of spermatogenesis since the germ cells enter meiosis around 9 dpp to form haploid spermatids at 20 dpp (Figure 1B). More than 30 000 piRNA sequences were identified from each of the complexes (Figure 1C and Supplementary information, Figure S1). Despite these large numbers of piRNAs identified, most of them in both populations were sequenced only once, indicating that the sequencing effort is still unsaturated for both. While this work was in progress, many more MILI- and MIWI-associated piRNAs were reported17,18.
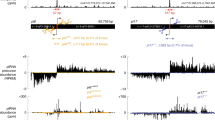
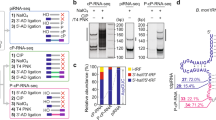
Postnatal testicular piRNAs are largely derived from non-transposon, intergenic regions. (A) MIWI and MILI associate with piRNAs in the testis. piRNAs isolated from the immunoprecipitates of MILI and MIWI were 5′ end-labeled and fractionated by 15% Urea-PAGE. Notice the size difference between MILI- and MIWI-associated piRNAs. MD: 10 nt DNA marker, which migrates 10% faster than the corresponding size of RNA. Adult testicular MILI immunoprecipitate (IP) data was used in a publication before30. (B) A diagram showing the expression of three mouse PIWI proteins during mouse testicular development. The bolts indicate the arrest points for the mutants. (C) Pie charts showing the number of the genomic hits of sequenced piRNAs from the MILI and MIWI complexes. MILI-associated piRNAs predominantly match to single (unique-mapping) sites on the genome throughout spermatogenesis as is the case for adult testicular MIWI-associated piRNAs. Repeat-associated piRNAs constitute a bigger fraction until 13 dpp relative to the adult testis. (D) Pie charts showing the genomic annotations of sequenced piRNAs from the MILI and MIWI complexes. MILI-associated piRNAs are largely derived from intergenic regions and poorly from transposon regions throughout spermatogenesis, as adult testicular MIWI-associated piRNAs. piRNAs from cellular RNAs and coding genes form a bigger fraction until 13 dpp relative to the adult stage. 13 dpp pachytene stage testis is rich in piRNAs that correspond to transposons relative to the other stages examined.
Since piRNAs involved in the ping-pong mechanism target complementary transposon intermediates and thus are abundant in transposon sequences19, we evaluated the genomic regions from which the cloned testicular piRNAs are derived. Mapping of these piRNAs on the genome revealed that majority of them is derived from unique sites, and repeat-associated sequences are underrepresented considering that such sequences constitute approximately 40% of the mouse genome (Figure 1C). Although relative to the adult testis, pre-meiotic testes of 7-8 dpp and early meiotic testes of 13 dpp are somewhat more abundant for piRNAs that have more than one mapping site, all four libraries are poor in transposon sequences (Figure 1D). Instead all these piRNA libraries are largely derived from non-transposon intergenic regions, implying that their biogenesis is mostly, if not completely, independent of the ping-pong machinery.
MILI- and MIWI-associated piRNAs do not contain the “ping-pong signature” or complementary sequences required for the ping-pong mechanism
To verify the above conclusion, we further examined whether MILI- and MIWI-associated piRNAs at 7-8 dpp, 13 dpp, and adult testes contain any ping-pong signature. piRNAs generated by the ping-pong machinery are complementary along their first 10 nucleotides due to the cleavage position of the “slicer” activity. Furthermore, since primary piRNAs are enriched with 5′ Uracil, ping-pong generates secondary piRNAs with Adenine bias on the 10th nucleotide position. We asked whether MILI- and MIWI- associated piRNA populations in the postnatal testes carry such a sequence signature. We found that they all are biased for Uracil on their 5′ ends, but neither piRNA population displays any nucleotide bias on the 10th nucleotide position (Figure 2A). Corroborating this observation, we found no significant enrichment of piRNAs with 10 nt complementarity between any pair of postnatal testicular libraries, even when we furthered our analysis to sub-populations of piRNAs derived from transposons and protein-coding genes (Figure 2B).
MILI- and MIWI-associated piRNAs do not contain “ping-pong signature” or complementary sequences required for the ping-pong mechanism. (A) Base composition of each piRNA population charted with the X-axis representing the nucleotide position relative to the 5′ (left panels) or 3′ (right panels) ends of the piRNAs (e.g., −1 at the 5′ end is the position 1-nt upstream of the piRNAs). The Y-axis represents the entropy score for the base bias. All populations of MILI-associated piRNAs are strongly biased for U on the 5′ end (position 1), and for G on the 2nd nucleotide position. In addition, they are biased for C on the 3′ end. MIWI-associated piRNAs show a bias for 5′ U only. None of the piRNA populations shows any A-bias at the 10th nucleotide position. (B) Pairwise comparison of the piRNA libraries shows that testicular piRNAs are largely devoid of matching piRNAs that are 10-nt complementary along their 5′ ends (left panel). The same has been observed for the transposon-derived sub-population of piRNAs (center panel), and for those derived from the coding genes (right panel). The values are the percentage of a given pair of piRNAs with complementary sequences, and belong to the libraries positioned on the rows. As a negative control, piRNA sequences from each library were randomized to generate a corresponding simulated library. Notice the similar percentage of complementary sequences in the libraries relative to their corresponding simulation.
MILI-associated piRNAs are upregulated but not downregulated in Miwi-null testes, in contrast to prediction by the ping-pong hypothesis
Next, we sought to experimentally address whether the ping-pong machinery is involved in the biogenesis of the adult testicular piRNAs. Since the ping-pong mechanism requires the interaction of two PIWI complexes carrying primary and secondary piRNAs, we first asked whether MILI and MIWI complexes interact with or depend on each other. For this purpose, we immunoprecipitated MILI to evaluate whether depleting the MILI complex would affect the MIWI complex. Analysis of their piRNA content by kinase labeling and gel electrophoresis showed that MILI and MIWI interact with distinct populations of piRNAs of different sizes in the same extract (Figure 3A), and the abundance of MIWI-associated piRNAs is not visibly affected by MILI immunoprecipitation. Likewise, immunoblotting of immunoprecipitates for MILI and MIWI showed no co-precipitation of MIWI or MILI. These observations indicate that MILI and MIWI complexes are not interdependent and do not physically interact with each other.
MILI-associated piRNAs are not downregulated in the absence of MIWI. (A) MILI and MIWI interact with independent populations of piRNAs. MILI and subsequently MIWI complexes were immunoprecipitated from the same adult testicular extract. Associated piRNAs were isolated and analyzed as in Figure 1. (B) Northern blotting for representative piRNAs. 20 μg of total testicular RNA samples from 24 dpp Miwi+/− and Miwi−/− were resolved with 15% Urea-PAGE and northern blots were probed for individual piRNAs. U6snRNA (U6) was used as an internal loading control. Ethidium bromide staining was also used to assess the global level of piRNAs. Five piRNA probes are used: Tp2, transposon 2; A-exo, anti-sense exonic; Rapi1, repeat-associated 1; S-Intro, sense intronic; T4, piRNA-T4. The annotations indicate the genomic regions from which piRNAs are derived (e.g., “anti-sense exonic” is a piRNA that corresponds to the anti-sense strand of an exon). Each probe hybridizes to both MIWI-associated (long arrows) and MILI-associated (short arrow) piRNAs. MILI-associated piRNAs are not downregulated in the absence of MIWI. (C) Immunoprecipitation of MILI from Miwi−/− testes, which was loaded with ∼1/3 of total RNAs as compared to that of Miwi+/− testes. Even though the non-piRNA controls (due to non-sepcific binding, empty arrow) are expectedly ∼1/3 lower in Miwi+/− testes, the total MILI-associated piRNAs in Miwi−/− testes (solid arrow) are equal to slightly more than that in Miwi+/− testes, indicating that the total MILI-associated piRNAs in increased by at least three times in Miwi−/− testes. IP, immunoprecipitates.
Since the ping-pong mechanism is a positive feedback loop between two different PIWI proteins for the production of each other's piRNAs, both piRNA populations produced by the mechanism should be drastically downregulated, if not eliminated, in the absence of either one of the proteins5,20,21. Therefore, we examined whether MILI-associated piRNAs are affected in the absence of MIWI by northern blotting for five representative MILI-associated piRNAs. Northern blotting for a MILI-associated piRNA typically yielded two bands with a strong one corresponding to the size of MIWI-associated piRNAs and a faint one with the size of MILI-associated piRNAs, indicating the close homology between the piRNA populations of MILI and MIWI. Our analyses show that surprisingly, none of the MILI-associated piRNAs is downregulated in the Miwi mutant (Figure 3B). In fact, they are all significantly upregulated in the Miwi mutant. We chose 24 dpp as the time point of the analysis because, in the absence of MIWI, spermatogenesis arrests at this point as early round spermatids. At 24 dpp, the Miwi−/− testis is identical in tissue composition to Miwi+/− at this stage14. Unfortunately we cannot conduct the same analysis for MIWI-associated piRNAs because the spermatogenesis in the Mili−/− mutant does not progress beyond the mid-pachytene stage and thus lacks the cells that express MIWI22.
To ascertain the small RNAs that we observe in Miwi−/− are indeed MILI-associated piRNAs but not de-stabilized MIWI-associated piRNAs, we performed β-elimination reaction to evaluate whether they are 2′-O-methylated at their 3′ termini. We deemed this analysis necessary since this modification is thought to maintain the stability of piRNAs23,24,25,26,27,28. In addition, the PIWI proteins in Drosophila interact with the piRNA methyltransferase, which in turn may be necessary for the methylation of piRNAs27. Noticeably, β-elimination reaction did not result a shift in the migration of the detected small RNAs in the Miwi−/− testis, in contrast to the internal miRNA control (Supplementary information, Figure S2A), indicating that they are modified at their 3′ termini. Furthermore, these small RNAs are depleted upon immunoprecipitation of MILI (Supplementary information, Figure S2B), confirming that they are MILI-associated piRNAs. Together, these observations confirm that they are intact piRNAs associated with MILI.
It is possible that MILI-associated piRNAs are overall downregulated while the particular ones that we tested are not. Therefore, we analyzed MILI-associated piRNAs at the global level by comparing the piRNA content of the MILI immunoprecipitates from 24 dpp Miwi−/− versus Miwi+/− testicular extract with end-labeling. Corroborating the northern analyses, we detected a significant increase in the level of MILI-associated piRNAs (Figure 3C), further supporting that piRNAs in the postnatal mouse testes are not produced by the ping-pong machinery.
MILI- and MIWI-associated piRNAs in the adult testis are produced by the primary pathway instead of the ping-pong mechanism
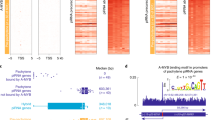
Then, what is the mode of piRNA biogenesis in postnatal testes? We asked whether we could deduce the mode of biogenesis based on the genomic regions from which the piRNAs are derived. It has been reported that murine piRNAs tend to be generated in a clustered fashion from specific loci in the genome9,10,11,12,13. However, it has not been shown whether postnatal MILI- and MIWI-associated piRNAs exhibit such a biogenesis pattern or whether they are produced from different loci. Our mapping of the piRNAs to the genome revealed that most of MILI- and MIWI-associated piRNAs are produced in a clustered fashion and, interestingly, from the same loci (Figure 4A and Supplementary information, Tables S1-S4). Furthermore, they are largely derived from the same loci at different stages of spermatogenesis (Supplementary information, Figure S3). Their genomic distribution within a cluster indicate that they are likely derived from the same precursor transcripts, as previously proposed, with some clusters containing only one transcript whereas others containing two transcripts divergently transcribed from a common promoter region, as previously reported10,12 (Figure 4B). Interestingly, we found that both MILI- and MIWI-associated piRNAs are derived from the same genomic strands within the clusters, which explains the lack of complementarity between the two populations and their close homology that we observed (Figure 4C). The common promoter region hypothesis for the bidirectionally-transcribed precursors in a cluster is supported by several observations: first, some long ESTs from the mouse testis correspond to the predicted piRNA precursors, including in the transcriptional direction and in the 5′ start positions (Figure 4C). Second, clustered piRNAs show head-to-tail homology reminiscent of their processing from the same type of precursors (Figure 4D). Third, the 3′ portion of precursors corresponds to piRNAs in a less dense fashion, which can be explained by a limited processivity of the RNA polymerase in transcribing the precursors (Figure 4B). Last, the promoter region is piRNA free (Figure 4C). Altogether, these bioinformatic analyses indicate that most postnatal MILI- and MIWI-associated piRNAs are derived from the same precursor transcripts via the primary piRNA biogenesis pathway instead of the ping-pong mechanism.
MILI- and MIWI-associated piRNAs in the adult testis are mostly processed from the same single-stranded precursor transcripts by the primary pathway instead of the ping-pong mechanism. (A) Mapping of the MILI- and MIWI-associated piRNAs reveals that they are both derived from the same sets of piRNA cluster loci. Cluster locations are indicated with triangles, whose height is proportional to the number of piRNAs in the cluster. The triangles on the left side of a chromosome denote piRNAs derived from the plus strand, while those on the right are from the minus strand. Blue and red triangles indicate MILI- and MIWI-associated piRNA clusters, respectively. Only clusters with more than 300 piRNAs are displayed. (B) piRNAs in clusters are derived from one or the other genomic strand. While piRNAs in a few clusters are derived from the same genomic strand along the entire cluster (bottom panel), in most clusters, they abruptly switch to the opposing strand at the center of the cluster resulting in a bidirectional profile (upper panel). Only MILI-associated piRNAs in the adult testis are shown. Each vertical bar represents one piRNA with the height of the bar reflecting the size of the corresponding piRNA. piRNAs from the plus strand are colored green, those from the minus strand are yellow. Arrows indicate the transcription direction of the piRNAs. (C) Higher resolution of the bidirectional cluster displayed in B shows that MILI- and MIWI-associated piRNAs are derived from the same genomic strands and switch origin of synthesis to the other strand at a same piRNA-free region. Only piRNAs cloned from the adult testicular immunoprecipitates are shown. EST and mRNA information from GenBank is displayed at the bottom. (D) Alignment of clustered piRNAs shows that they have overlapping sequences. Shown is part of the genomic sequence (top line) of the Chromosome 17 cluster and the piRNAs that correspond to this sequence.
Transposon-derived piRNAs are processed from precursors but not by the ping-pong mechanism
Although a minor fraction of transposon piRNAs exist in the three libraries analyzed, they seems to be a result of their residence in the piRNA cluster loci, where they are processed as part of the putative piRNA precursors rather than through a ping-pong mechanism (Figure 5). On the other hand, when intron-containing sequences serve as piRNA precursors or a part of a larger piRNA precursor, they tend to undergo splicing before they are cleaved into piRNAs, as evidenced by our observation that unique intron-derived piRNAs are underrepresented (Supplementary information, Figure S4).
Discussion
In this paper, we have provided definitive evidence indicating that piRNAs in postnatal testes are generated mostly, if not exclusively, by the primary pathway but not ping-pong pathway. This is in contrast to the common belief that the ping-pong pathway is a major mechanism for piRNA biogenesis during spermatogenesis. Furthermore, our results also reveal a possible competition between MILI and MIWI for the same precursors of their piRNAs. This hypothesis is supported by two clear lines of evidence: first, MILI and MIWI share common piRNA precursors; and second, all of five piRNAs that were subjected to northern analysis, as well as total MILI-associated piRNAs as examined by immunoprecipitation experiments of MILI, are significantly upregulated in the absence of MIWI (Figure 3). This may be a unique feature of the primary processing mechanism involved in piRNA biogenesis.
We also noticed that MILI-associated piRNAs at the pachytene stage (13 dpp) are relatively rich in sequences derived from protein-coding genes. This stage corresponds to an overall increase in transcriptional activity29. It is possible that some of the resultant mRNAs might somehow enter the piRNA biogenesis pathway to be processed into the observed gene-derived piRNAs. Supporting this view, most of these piRNAs correspond to the sense strand of the genes. Our analysis reported here points to the importance of elucidating the primary processing mechanism, which at present received little attention, especially compared to the ping-pong mechanism, and thus remains largely elusive.
Materials and Methods
Mouse strains
The generation and genetic backgrounds of Mili−/−22 and Miwi−/−14 mice are described previously. The homozygous mutants were obtained by crossing heterozygous males to homozygous mutant females. CD1 strain mice were used as the wild-type model and in the identification of the piRNAs associated with MILI and MIWI.
Antibodies
MIWI and MILI complexes were immunoprecipitated with the R133 anti-MIWI serum14, and affinity-purified MILI peptide antibody30, respectively.
Immunoprecipitation of the PIWI/piRNA complexes
Immunoprecipitation was performed as described in30 from the crude lysates of mouse testes with the following modifications: briefly, mice of the indicated ages were euthanized with cervical dislocation or asphyxiation with CO2. Testes were dissected out, flash frozen in liquid nitrogen and stored at −80 °C until needed. After removing tunica albuginea, they were homogenized in 50 mM HEPES pH 7.4, 150 mM KOAc, 0.1% Triton X-100, 0.1% NP-40, 2 mM MgOAc, 10% glycerol, 1 mM DTT, 80 μ/ml RNaseOUT (Invitrogen), complete mini EDTA-free protease inhibitor cocktail tablet (Roche), 200 μM Cycloheximide with a dounce homogenizer for at least 20 strokes under RNase-free conditions. Immunoprecipitations were performed overnight at 4 °C using protein A-Sepharose beads conjugated with affinity-purified MILI peptide antibody or R133 anti-MIWI serum. As the negative controls, antibodies blocked with the antigen peptide as in30 and antibody-free protein A-Sepharose beads were used for the MILI and MIWI immunoprecipitations, respectively.
piRNA cloning and sequencing
For the identification of the piRNAs, co-immunoprecipitated RNA was extracted from the immunoprecipitates with TRIZOL Reagent (Invitrogen), and 5′ end-labeled with [32P-γ] ATP by T4 Polynucleotide Kinase (New England Biolabs). Following the labeling, the sample was saturated with non-radioactive ATP to ensure all the 5′ ends are phosphorylated, which is required to ligate the 5′ adapter during the cloning. The samples were purified off un-incorporated nucleotides with Sephadex G-25 columns (Amersham Biosciences) and resolved with 10% Urea-PAGE alongside end-labeled 10-nucleotide DNA marker (Invitrogen) to distinguish the co-immunoprecipitated piRNA population. Under the denaturing conditions that we employed, the DNA marker migrates 10% faster than RNA. piRNAs were extracted from the gel and precipitated with NaOAce/EtOH. Cloning was performed as in31. The adapters were obtained from Integrated DNA Technologies (IDT). cDNAs of the piRNAs were amplified with PCR and the resulting pi-DNA libraries were identified with high throughput pyro-sequencing by 454 (Roche).
Denaturing PAGE analysis of piRNAs
The indicated amount of RNA samples were denatured in 50% formamide at 55 °C for 15 min. The samples were resolved in polyacrylamide gel containing 6 M Urea with 1× TBE. For the analysis of the radiolabeled samples, the gels were exposed to X-ray films (Kodak) or phosphorimager screen.
Small RNA northern blotting
Following denaturing PAGE, the gel was stained with 1 μg/ml ethidium bromide in 1× TBE for ∼15 min to assess the global piRNA content and integrity of the samples. Afterwards, the gel was de-stained in 1× TBE for ∼10 min and transferred onto Hybond-N nylon membrane (Amersham Biosciences) in 1× TBE for 30 min at 350 milliAmp using Hoefer TE 22 tank transfer unit at 4 °C. The membrane was dried at 75 °C for 5-10 min. The samples were cross-linked to the membrane with UV light of 120 mJ/cm2 followed by baking at 75 °C for 1 h. The membrane was stored at −80 °C until needed or used immediately for probing. Hybridizations were performed overnight at 42 °C in 5× SSC, 20 mM Na2HPO4 at pH 7.2, 7% SDS, 1× Denhardt's solution, 0.1 mg/ml boiled salmon sperm DNA following pre-hybridization in the same buffer composition without Denhardt's Solution for at least 30 min at 42 °C. Probes were prepared as follows: DNA oligonucleotides with reverse complementary sequences for individual small RNAs were obtained from IDT and radiolabeled on their 5′ ends with kinase reaction. For piRNAs, oligos were LNA modified to yield a Tm value of 75 °C (±3 °C). Labeled probes were boiled for 1 min before adding into the hybridization buffer. Following hybridization, blots were washed twice in 1× SSC and 0.1% SDS at 42 °C for 10 min and analyzed by PhosphorImager. If necessary, blots were stripped by boiling in 0.1% SDS for 10 min.
Analysis of the 3′ ends of the piRNAs in the Miwi−/− testis
Modification status of the piRNA 3′ termini was determined with β-elimination reaction as in26. 40 μg of total RNA from a group of 23-24 dpp Miwi−/− testes alongside 23-24 dpp Miwi+/− testes was subjected to periodate oxidation and β-elimination. Samples were resolved with 20% urea-PAGE and analyzed with northern blotting. Controls with no β-elimination were performed alongside experimental samples by substituting H2O for NaIO4, borax-boric acid buffer and NaOH. miRNA-16, which is not modified on its 3′ end, was used as a positive control for the β-elimination reaction.
Probes used for northern blotting
The nucleotides with the “+” sign on their left are LNA-modified.
mir-16: CGC CAA TAT TTA CGT GCT GCT A
U6 snRNA: TGT GCT GCC GAA GCG AGC AC
piRNA probes:
piRNA T4 is a MIWI-associated piRNA, the rest are MILI-associated.
Transposonic 2: GGA C+CG+GT+C T+GC AG+C TGC TGA GTC GTA
Anti-sense exonic: GGG ACA CA+C T+CA+GC+A C+TC+CT+T TGC A
Sense intronic 1: GGG CAG GTG+AG+A G+GA+TCC ATG GCC CA
Repeat-associated 1: TA+T CA+T A+GT +CA+T CA+T CA+T C+AT +CG+T CA
piRNA T4: TAG ACA ATT TTC AGT GTC CTA AGC TGT CTA
Preliminary bioinformatic processing of small RNA libraries
Cloned small RNA sequences were first clipped off the linkers. We were able to clip more than 92% of the sequences in each library (Supplementary information, Figure S1A). Over 80% of the total small RNAs fell into the clipped clones of 18-35 nt size range in the MILI immunoprecipitates, and 20-39 nt size range in the MIWI immunoprecipitates. These piRNAs were defined as MILI- and MIWI-associated piRNAs, respectively. Following this classification, piRNAs were mapped to the mouse genome (mm9) for further analysis allowing up to 2 mismatches, which includes insertions, deletions and substitutions. More than 72% of the total cloned sequences could be mapped on the genome in each library.
Small RNA annotation and size profile analysis
Small RNAs that can be mapped to rRNA, tRNA, snoRNA, snRNA were annotated and defined as “cellular RNAs”. mirBase depository was used for microRNA annotation. The annotation information from RepeatMasker was referred for transposon annotation. As for the gene annotation, Ensembl database was used. Size profile of the small RNAs was evaluated with the small RNA abundance normalized to the sequencing depth in each library (Supplementary information, Figure S1B).
Small RNA sequence feature and ping-pong signature analysis
piRNAs were analyzed for their overall base composition in their first 10 nt (5′ end) and in their last 10 nt (3′ end), as well as in their surrounding sequences. The cloning frequency of the piRNAs was integrated into the analysis. piRNA partners that are complimentary to each other on their 5′ ends (10 nt off-set) among the libraries were determined as previously reported2,3,4,5,6,7. As a negative control, a simulation was performed with the randomized sequences that were created based on the frequencies of the bases in the libraries. From 7-8 dpp to adult stage, MILI-bound piRNAs are always biased to 24-28 nucleotides long. Meanwhile, we noticed that MILI 7-8 dpp piRNAs have broader range in size compared to 13 dpp and adult stages. Comparatively, adult MIWI-bound piRNAs are biased to 29-31 nucleotides in size.
piRNA cluster analysis
A piRNA cluster was defined as a group of piRNAs with at least 50 piRNAs, where the piRNAs are less than 1 500 bp away from each other. Each clone that maps to only one location (defined as “unique”) on the genome was counted as one piRNA, while those that map to x number of locations were counted as 1/x piRNA in the estimation of the piRNA amount in a cluster with the exception in Figure 5, where every clone was counted as 1 piRNA regardless of the number of regions that they map to. The number of piRNAs per cluster has also been normalized based on the sequencing coverage of the library. MILI-associated piRNAs of 7-8 dpp, 13 dpp and adult testes yielded 16, 22 and 21 clusters, respectively, and 25 clusters were estimated for the MIWI-associated piRNAs in the adult. These clustered piRNAs cover 46%, 51%, 71% and 84% of the piRNAs in the libraries of MILI7-8 dpp, MILI13 dpp, MILI-adult and MIWI-adult immunoprecipitates, respectively.
References
Juliano C, Wang J, Lin H . Uniting Germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet 2011; 45:447–469.
Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007; 128:1089–1103.
Gunawardane LS, Saito K, Nishida KM, et al. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 2007; 315:1587–1590.
Houwing S, Berezikov E, Ketting RF . Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 2008; 27:2702–2711.
Aravin AA, Sachidanandam R, Bourc'his D, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 2008; 31:785–799.
Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ . Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 2007; 316:744–747.
Klattenhoff C, Theurkauf W . Biogenesis and germline functions of piRNAs. Development 2008; 135:3–9.
Kuramochi-Miyagawa S, Watanabe T, Gotoh K, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 2008; 22:908–917.
Lau NC, Seto AG, Kim J, et al. Characterization of the piRNA complex from rat testes. Science 2006; 313:363–367.
Girard A, Sachidanandam R, Hannon GJ, Carmell MA . A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006; 442:199–202.
Watanabe T, Takeda A, Tsukiyama T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 2006; 20:1732–1743.
Aravin A, Gaidatzis D, Pfeffer S, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006; 442:203–207.
Grivna ST, Beyret E, Wang Z, Lin H . A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 2006; 20:1709–1714.
Deng W, Lin H . miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2002; 2:819–830.
Wang J, Saxe JP, Tanaka T, Chuma S, Lin H . Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol 2009; 19:640–644.
Kuramochi-Miyagawa S, Kimura T, Yomogida K, et al. Two mouse piwi-related genes: miwi and mili. Mech Dev 2001; 108:121–133.
Reuter M, Chuma S, Tanaka T, et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 2009; 16:639–646.
Robine N, Lau NC, Balla S, et al. A broadly conserved pathway generates 3'UTR-directed primary piRNAs. Curr Biol 2009; 19:2066–2076.
Thomson T, Lin H . The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 2009; 25:355–376.
Li C, Vagin VV, Lee S, et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 2009; 137:509–521.
Malone CD, Brennecke J, Dus M, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009; 137:522–535.
Kuramochi-Miyagawa S, Kimura T, Ijiri TW, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 2004; 131:839–849.
Kirino Y, Mourelatos Z . Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 2007; 14:347–348.
Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T . The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 2007; 14:349–350.
Houwing S, Kamminga LM, Berezikov E, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 2007; 129:69–82.
Horwich MD, Li C, Matranga C, et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 2007; 17:1265–1272.
Saito K, Sakaguchi Y, Suzuki T, et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi-interacting RNAs at their 3′ ends. Genes Dev 2007; 21:1603–1608.
Kurth HM, Mochizuki K . 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA 2009; 15:675–685.
Bellve AR . The molecular biology of mammalian spermatogenesis. In: Finn CA, ed. Oxford Review of Reproductive Biology. Oxford: Oxford University Clarendon Press, 1979:159–261.
Unhavaithaya Y, Hao Y, Beyret E, et al. MILI, a piRNA binding protein, is required for germline stem cell self-renewal and appears to positively regulate translation. J Biol Chem 2008; 284:6507–6519.
Lau NC, Lim LP, Weinstein EG, Bartel DP . An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001; 294:858–862.
Acknowledgements
We thank Dr Hang Yin (Yale University) for initial bioinformatics analysis. This work is supported by National Institutes of Health (HD42012), and the G. Harold and Leila Mathers Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
( Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary information
Supplementary information, Figure S1
Overall profiles of MILI- and MIWI-associated piRNAs. (PDF 142 kb)
Supplementary information, Figure S2
piRNAs observed in the Miwi−/− testis are intact MILI-associated piRNAs. (PDF 119 kb)
Supplementary information, Figure S3
piRNAs are produced from common hot spots on the genome during spermatogenesis. (PDF 96 kb)
Supplementary information, Figure S4
Profile of piRNAs corresponding to protein-coding genes. (PDF 157 kb)
Supplementary information, Table S1
Genomic Loci of the MILI-associated piRNA clusters in the 7-8dpp testis. (PDF 54 kb)
Supplementary information, Table S2
Genomic Loci of the MILI-associated piRNA clusters in the 13dpp testis. (PDF 57 kb)
Supplementary information, Table S3
Genomic Loci of the MILI-associated piRNA clusters in the adult testis. (PDF 53 kb)
Supplementary information, Table S4
Genomic Loci of the MIWI-associated piRNA clusters in the adult testis. (PDF 55 kb)
Rights and permissions
About this article
Cite this article
Beyret, E., Liu, N. & Lin, H. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res 22, 1429–1439 (2012). https://doi.org/10.1038/cr.2012.120
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cr.2012.120
Keywords
This article is cited by
-
Emerging roles and functional mechanisms of PIWI-interacting RNAs
Nature Reviews Molecular Cell Biology (2023)
-
PiRNA in Cardiovascular Disease: Focus on Cardiac Remodeling and Cardiac Protection
Journal of Cardiovascular Translational Research (2023)
-
An evolutionarily conserved stop codon enrichment at the 5′ ends of mammalian piRNAs
Nature Communications (2022)
-
piRNAs and PIWI proteins as potential biomarkers in Breast cancer
Molecular Biology Reports (2022)
-
The birth of piRNAs: how mammalian piRNAs are produced, originated, and evolved
Mammalian Genome (2022)