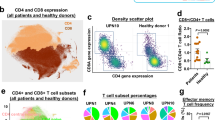
Abstract
Immunoglobulin (IG) gene repertoire restrictions strongly support antigen selection in the pathogenesis of chronic lymphocytic leukemia (CLL). Given the emerging multifarious interactions between CLL and bystander T cells, we sought to determine whether antigen(s) are also selecting T cells in CLL. We performed a large-scale, next-generation sequencing (NGS) study of the T-cell repertoire, focusing on major stereotyped subsets representing CLL subgroups with undisputed antigenic drive, but also included patients carrying non-subset IG rearrangements to seek for T-cell immunogenetic signatures ubiquitous in CLL. Considering the inherent limitations of NGS, we deployed bioinformatics algorithms for qualitative curation of T-cell receptor rearrangements, and included multiple types of controls. Overall, we document the clonal architecture of the T-cell repertoire in CLL. These T-cell clones persist and further expand overtime, and can be shared by different patients, most especially patients belonging to the same stereotyped subset. Notably, these shared clonotypes appear to be disease-specific, as they are found in neither public databases nor healthy controls. Altogether, these findings indicate that antigen drive likely underlies T-cell expansions in CLL and may be acting in a CLL subset-specific context. Whether these are the same antigens interacting with the malignant clone or tumor-derived antigens remains to be elucidated.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Stamatopoulos K, Belessi C, Moreno C, Boudjograh M, Guida G, Smilevska T et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood 2007; 109: 259–270.
Baliakas P, Hadzidimitriou A, Sutton LA, Minga E, Agathangelidis A, Nichelatti M et al. Clinical effect of stereotyped B-cell receptor immunoglobulins in chronic lymphocytic leukaemia: a retrospective multicentre study. Lancet Haematol 2014; 1: e74–e84.
Tobin G, Thunberg U, Johnson A, Eriksson I, Soderberg O, Karlsson K et al. Chronic lymphocytic leukemias utilizing the VH3-21 gene display highly restricted Vlambda2-14 gene use and homologous CDR3s: implicating recognition of a common antigen epitope. Blood 2003; 101: 4952–4957.
Agathangelidis A, Darzentas N, Hadzidimitriou A, Brochet X, Murray F, Yan XJ et al. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood 2012; 119: 4467–4475.
Baliakas P, Agathangelidis A, Hadzidimitriou A, Sutton LA, Minga E, Tsanousa A et al. Not all IGHV3-21 chronic lymphocytic leukemias are equal: prognostic considerations. Blood 2015; 125: 856–859.
Gounari M, Ntoufa S, Apollonio B, Papakonstantinou N, Ponzoni M, Chu CC et al. Excessive antigen reactivity may underlie the clinical aggressiveness of chronic lymphocytic leukemia stereotyped subset #8. Blood 2015; 125: 3580–3587.
Burger JA, Tedeschi A, Barr PM, Robak T, Owen C, Ghia P et al. Ibrutinib as initial therapy for patients with chronic lymphocytic leukemia. N Engl J Med 2015; 373: 2425–2437.
Brown JR, Barrientos JC, Barr PM, Flinn IW, Burger JA, Tran A et al. The Bruton tyrosine kinase inhibitor ibrutinib with chemoimmunotherapy in patients with chronic lymphocytic leukemia. Blood 2015; 125: 2915–2922.
Brown JR, Byrd JC, Coutre SE, Benson DM, Flinn IW, Wagner-Johnston ND et al. Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110delta, for relapsed/refractory chronic lymphocytic leukemia. Blood 2014; 123: 3390–3397.
Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med 2013; 369: 32–42.
Burger JA, Gribben JG . The microenvironment in chronic lymphocytic leukemia (CLL) and other B cell malignancies: insight into disease biology and new targeted therapies. Semin Cancer Biol 2014; 24: 71–81.
Bagnara D, Kaufman MS, Calissano C, Marsilio S, Patten PE, Simone R et al. A novel adoptive transfer model of chronic lymphocytic leukemia suggests a key role for T lymphocytes in the disease. Blood 2011; 117: 5463–5472.
Gorgun G, Holderried TA, Zahrieh D, Neuberg D, Gribben JG . Chronic lymphocytic leukemia cells induce changes in gene expression of CD4 and CD8 T cells. J Clin Invest 2005; 115: 1797–1805.
Os A, Burgler S, Ribes AP, Funderud A, Wang D, Thompson KM et al. Chronic lymphocytic leukemia cells are activated and proliferate in response to specific T helper cells. Cell Rep 2013; 4: 566–577.
Ramsay AG, Johnson AJ, Lee AM, Gorgun G, Le Dieu R, Blum W et al. Chronic lymphocytic leukemia T cells show impaired immunological synapse formation that can be reversed with an immunomodulating drug. J Clin Invest. 2008; 118: 2427–2437.
Ramsay AG, Clear AJ, Fatah R, Gribben JG . Multiple inhibitory ligands induce impaired T-cell immunologic synapse function in chronic lymphocytic leukemia that can be blocked with lenalidomide: establishing a reversible immune evasion mechanism in human cancer. Blood 2012; 120: 1412–1421.
Ramsay AG, Evans R, Kiaii S, Svensson L, Hogg N, Gribben JG . Chronic lymphocytic leukemia cells induce defective LFA-1-directed T-cell motility by altering Rho GTPase signaling that is reversible with lenalidomide. Blood 2013; 121: 2704–2714.
Shanafelt TD, Ramsay AG, Zent CS, Leis JF, Tun HW, Call TG et al. Long-term repair of T-cell synapse activity in a phase II trial of chemoimmunotherapy followed by lenalidomide consolidation in previously untreated chronic lymphocytic leukemia (CLL). Blood 2013; 121: 4137–4141.
Vardi A, Agathangelidis A, Stalika E, Karypidou M, Siorenta A, Anagnostopoulos A et al. Antigen Selection shapes the t-cell repertoire in chronic lymphocytic leukemia. Clin Cancer Res 2016; 22: 167–174.
van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 concerted action BMH4-CT98-3936. Leukemia 2003; 17: 2257–2317.
Venturi V, Kedzierska K, Price DA, Doherty PC, Douek DC, Turner SJ et al. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc Natl Acad Sci USA 2006; 103: 18691–18696.
Quigley MF, Greenaway HY, Venturi V, Lindsay R, Quinn KM, Seder RA et al. Convergent recombination shapes the clonotypic landscape of the naive T-cell repertoire. Proc Natl Acad Sci USA 2010; 107: 19414–19419.
Sutton LA, Kostareli E, Hadzidimitriou A, Darzentas N, Tsaftaris A, Anagnostopoulos A et al. Extensive intraclonal diversification in a subgroup of chronic lymphocytic leukemia patients with stereotyped IGHV4-34 receptors: implications for ongoing interactions with antigen. Blood 2009; 114: 4460–4468.
Kostareli E, Sutton LA, Hadzidimitriou A, Darzentas N, Kouvatsi A, Tsaftaris A et al. Intraclonal diversification of immunoglobulin light chains in a subset of chronic lymphocytic leukemia alludes to antigen-driven clonal evolution. Leukemia 2010; 24: 1317–1324.
Sutton LA, Kostareli E, Stalika E, Tsaftaris A, Anagnostopoulos A, Darzentas N et al. Temporal dynamics of clonal evolution in chronic lymphocytic leukemia with stereotyped IGHV4-34/IGKV2-30 antigen receptors: longitudinal immunogenetic evidence. Mol Med 2013; 19: 230–236.
Sutton LA, Rosenquist R . Clonal evolution in chronic lymphocytic leukemia: impact of subclonality on disease progression. Expert Rev Hematol 2015; 8: 71–78.
Murray F, Darzentas N, Hadzidimitriou A, Tobin G, Boudjogra M, Scielzo C et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood 2008; 111: 1524–1533.
Chu CC, Catera R, Zhang L, Didier S, Agagnina BM, Damle RN et al. Many chronic lymphocytic leukemia antibodies recognize apoptotic cells with exposed nonmuscle myosin heavy chain IIA: implications for patient outcome and cell of origin. Blood 2010; 115: 3907–3915.
Duhren-von Minden M, Ubelhart R, Schneider D, Wossning T, Bach MP, Buchner M et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 2012; 489: 309–312.
Gounari M, Minici C, Ubelhart R, Duehren-von Minden M, Schneider D, Tasdogan A et al. Distinct homotypic B-cell receptor interactions shape the outcome of chronic lymphocytic leukemia. 21st Congress of EHA; 2016. Copenhagen, Denmark: Haematologica, 2016.
McClanahan F, Hanna B, Miller S, Clear AJ, Lichter P, Gribben JG et al. PD-L1 checkpoint blockade prevents immune dysfunction and leukemia development in a mouse model of chronic lymphocytic leukemia. Blood 2015; 126: 203–211.
McClanahan F, Riches JC, Miller S, Day WP, Kotsiou E, Neuberg D et al. Mechanisms of PD-L1/PD-1–mediated CD8 T-cell dysfunction in the context of aging-related immune defects in the Eμ-TCL1 CLL mouse model. Blood 2015; 126: 212–221.
Acknowledgements
Supported in part by H2020 ‘AEGLE, An analytics framework for integrated and personalized healthcare services in Europe’, by the EU; ‘MEDGENET, Medical Genomics and Epigenomics Network’ (No.692298) by the EU. Part of the work was carried out with the support of core facilities of CEITEC – Central European Institute of Technology under CEITEC – open access project, ID number LM2011020, funded by the Ministry of Education, Youth and Sports of the Czech Republic under the activity ‘Projects of major infrastructures for research, development and innovations’. PS and BV are financially supported by the Ministry of Education, Youth and Sports of the Czech Republic under the project CEITEC 2020 (LQ1601) and by the Horizon2020 Programme Twinning (MEDGENET/2016–2018/no.692298).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Vardi, A., Vlachonikola, E., Karypidou, M. et al. Restrictions in the T-cell repertoire of chronic lymphocytic leukemia: high-throughput immunoprofiling supports selection by shared antigenic elements. Leukemia 31, 1555–1561 (2017). https://doi.org/10.1038/leu.2016.362
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.362
This article is cited by
-
Activation and expansion of T-follicular helper cells in chronic lymphocytic leukemia nurselike cell co-cultures
Leukemia (2022)
-
TRIP - T cell receptor/immunoglobulin profiler
BMC Bioinformatics (2020)
-
T-cells in chronic lymphocytic leukemia: Guardians or drivers of disease?
Leukemia (2020)
-
Control of chronic lymphocytic leukemia development by clonally-expanded CD8+ T-cells that undergo functional exhaustion in secondary lymphoid tissues
Leukemia (2019)
-
Profiling the B/T cell receptor repertoire of lymphocyte derived cell lines
BMC Cancer (2018)