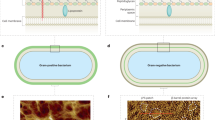
Abstract
An important goal of membrane biology is to define the local heterogeneity of membrane lipid composition. Here we describe a quantitative electron microscopic method that enables the localization of specific membrane lipids at the nanometer scale. The method involves freezing cells rapidly to halt the molecular motion, physically stabilizing membrane molecules in the freeze-fracture replica by the deposition of evaporated platinum and carbon layers and labeling with specific probes for electron microscopic observation. Lipids in both the outer and inner membrane leaflets can thus be labeled, and their distributions can be analyzed quantitatively by statistical methods. A major advantage of this method is that it does not require the expression of artificial probes. Therefore, this method can be applied to any cell in vitro or in vivo, and the whole procedure can be completed in 1–2 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Di Paolo, G. & De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 (2006).
Edidin, M. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 32, 257–283 (2003).
Hancock, J.F. Lipid rafts: contentious only from simplistic standpoints. Nat. Rev. Mol. Cell. Biol. 7, 456–462 (2006).
Simons, K. & Vaz, W.L. Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33, 269–295 (2004).
Stauffer, T.P., Ahn, S. & Meyer, T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8, 343–346 (1998).
Varnai, P. & Balla, T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell. Biol. 143, 501–510 (1998).
Downes, C.P., Gray, A. & Lucocq, J.M. Probing phosphoinositide functions in signaling and membrane trafficking. Trends. Cell. Biol. 15, 259–268 (2005).
Irvine, R. Inositol lipids: to PHix or not to PHix? Curr. Biol. 14, R308–R310 (2004).
Kusumi, A. & Suzuki, K. Toward understanding the dynamics of membrane-raft-based molecular interactions. Biochim. Biophys. Acta. 1746, 234–251 (2005).
Heuser, J.E. et al. Synaptic vesicle exocytosis captured by quick freezing and correlated with quantal transmitter release. J. Cell. Biol. 81, 275–300 (1979).
Dahl, R. & Staehelin, L.A. High-pressure freezing for the preservation of biological structure: theory and practice. J. Electron. Microsc. Tech. 13, 165–174 (1989).
Fujimoto, K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J. Cell. Sci. 108, 3443–3449 (1995).
Fujimoto, K., Umeda, M. & Fujimoto, T. Transmembrane phospholipid distribution revealed by freeze-fracture replica labeling. J. Cell. Sci. 109, 2453–2460 (1996).
Fujita, A. & Fujimoto, T. Quantitative retention of membrane lipids in the freeze-fracture replica. Histochem. Cell. Biol. 128, 385–389 (2007).
Fujita, A. et al. Gangliosides GM1 and GM3 in the living cell membrane form clusters susceptible to cholesterol depletion and chilling. Mol. Biol. Cell. 18, 2112–2122 (2007).
Fujita, A., Cheng, J. & Fujimoto, T. Segretation of GM1 and GM3 clusters in the cell membrane depends on the intact actin cytoskeleton. Biochim. Biophys. Acta. 1791, 388–396 (2009).
Fujita, A., Cheng, J., Tauchi-Sato, K., Takenawa, T. & Fujimoto, T. A distinct pool of phosphatidylinositol 4,5-bisphosphate in caveolae revealed by a nanoscale labeling technique. Proc. Natl. Acad. Sci. USA 106, 9256–9261 (2009).
Prior, I.A., Muncke, C., Parton, R.G. & Hancock, J.F. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell. Biol. 160, 165–170 (2003).
Vanhecke, D., Graber, W. & Studer, D. Close-to-native ultrastructural preservation by high pressure freezing. Methods. Cell. Biol. 88, 151–164 (2008).
Hagiwara, A., Fukazawa, Y., Deguchi-Tawarada, M., Ohtsuka, T. & Shigemoto, R. Differential distribution of release-related proteins in the hippocampal CA3 area as revealed by freeze-fracture replica labeling. J. Comp. Neurol. 489, 195–216 (2005).
Ripley, B.D. Tests of randomness for spatial point patterns. J. R. Stat. Soc. Ser. B 41, 368–374 (1979).
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research and the Global COE Program 'Integrated Molecular Medicine for Neuronal and Neoplastic Disorders' of the Ministry of Education, Culture, Sports, Science and Technology of the Japanese Government.
Author information
Authors and Affiliations
Contributions
A.F. performed the experiments and prepared the manuscript. J.C. technically assisted. T.F. designed the experiments and prepared the manuscript.
Corresponding author
Rights and permissions
About this article
Cite this article
Fujita, A., Cheng, J. & Fujimoto, T. Quantitative electron microscopy for the nanoscale analysis of membrane lipid distribution. Nat Protoc 5, 661–669 (2010). https://doi.org/10.1038/nprot.2010.20
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.20
This article is cited by
-
Ceramide structure dictates glycosphingolipid nanodomain assembly and function
Nature Communications (2021)
-
Glycosphingolipid GM3 is localized in both exoplasmic and cytoplasmic leaflets of Plasmodium falciparum malaria parasite plasma membrane
Scientific Reports (2021)
-
The distribution of phosphatidylinositol 4,5-bisphosphate in the budding yeast plasma membrane
Histochemistry and Cell Biology (2021)
-
Nanoscale analysis reveals no domain formation of glycosylphosphatidylinositol-anchored protein SAG1 in the plasma membrane of living Toxoplasma gondii
Histochemistry and Cell Biology (2019)
-
Measurement of caveolin-1 densities in the cell membrane for quantification of caveolar deformation after exposure to hypotonic membrane tension
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.