Abstract
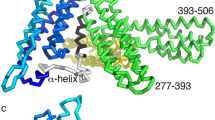
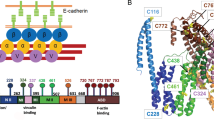
The F-actin–binding cytoskeletal protein α-catenin interacts with β-catenin–cadherin complexes and stabilizes cell-cell junctions. The β-catenin–α-catenin complex cannot bind F-actin, whereas interactions of α-catenin with the cytoskeletal protein vinculin appear to be necessary to stabilize adherens junctions. Here we report the crystal structure of nearly full-length human α-catenin at 3.7-Å resolution. α-catenin forms an asymmetric dimer where the four-helix bundle domains of each subunit engage in distinct intermolecular interactions. This results in a left handshake–like dimer, wherein the two subunits have remarkably different conformations. The crystal structure explains why dimeric α-catenin has a higher affinity for F-actin than does monomeric α-catenin, why the β-catenin–α-catenin complex does not bind F-actin, how activated vinculin links the cadherin–catenin complex to the cytoskeleton and why α-catenin but not inactive vinculin can bind F-actin.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Volk, T. & Geiger, B. A 135-kd membrane protein of intercellular adherens junctions. EMBO J. 3, 2249–2260 (1984).
Takeichi, M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development 102, 639–655 (1988).
Nishimura, T. & Takeichi, M. Remodeling of the adherens junctions during morphogenesis. Curr. Top. Dev. Biol. 89, 33–54 (2009).
Brasch, J., Harrison, O.J., Honig, B. & Shapiro, L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 22, 299–310 (2012).
Hirano, S., Nose, A., Hatta, K., Kawakami, A. & Takeichi, M. Calcium-dependent cell-cell adhesion molecules (cadherins): subclass specificities and possible involvement of actin bundles. J. Cell Biol. 105, 2501–2510 (1987).
Takeichi, M. Morphogenetic roles of classic cadherins. Curr. Opin. Cell Biol. 7, 619–627 (1995).
Pokutta, S. & Weis, W.I. Structure and mechanism of cadherins and catenins in cell-cell contacts. Annu. Rev. Cell Dev. Biol. 23, 237–261 (2007).
Lecuit, T. α-catenin mechanosensing for adherens junctions. Nat. Cell Biol. 12, 522–524 (2010).
Miyake, Y. et al. Actomyosin tension is required for correct recruitment of adherens junction components and zonula occludens formation. Exp. Cell Res. 312, 1637–1650 (2006).
Hirano, S., Kimoto, N., Shimoyama, Y., Hirohashi, S. & Takeichi, M. Identification of a neural α-catenin as a key regulator of cadherin function and multicellular organization. Cell 70, 293–301 (1992).
Watabe, M., Nagafuchi, A., Tsukita, S. & Takeichi, M. Induction of polarized cell-cell association and retardation of growth by activation of the E-cadherin–catenin adhesion system in a dispersed carcinoma line. J. Cell Biol. 127, 247–256 (1994).
Torres, M. et al. An α-E-catenin gene trap mutation defines its function in preimplantation development. Proc. Natl. Acad. Sci. USA 94, 901–906 (1997).
Watabe-Uchida, M. et al. α-Catenin–vinculin interaction functions to organize the apical junctional complex in epithelial cells. J. Cell Biol. 142, 847–857 (1998).
Vasioukhin, V., Bauer, C., Degenstein, L., Wise, B. & Fuchs, E. Hyperproliferation and defects in epithelial polarity upon conditional ablation of α-catenin in skin. Cell 104, 605–617 (2001).
Drees, F., Pokutta, S., Yamada, S., Nelson, W.J. & Weis, W.I. α-catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell 123, 903–915 (2005).
Weis, W.I. & Nelson, W.J. Re-solving the cadherin-catenin-actin conundrum. J. Biol. Chem. 281, 35593–35597 (2006).
Yamada, S., Pokutta, S., Drees, F., Weis, W.I. & Nelson, W.J. Deconstructing the cadherin-catenin-actin complex. Cell 123, 889–901 (2005).
Peng, X., Cuff, L.E., Lawton, C.D. & DeMali, K.A. Vinculin regulates cell-surface E-cadherin expression by binding to β-catenin. J. Cell Sci. 123, 567–577 (2010).
Yonemura, S., Wada, Y., Watanabe, T., Nagafuchi, A. & Shibata, M. α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 (2010).
Rangarajan, E.S. & Izard, T. α-Catenin unfurls upon binding to vinculin. J. Biol. Chem. 287, 18492–18499 (2012).
Abe, K. & Takeichi, M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc. Natl. Acad. Sci. USA 105, 13–19 (2008).
Hazan, R.B., Kang, L., Roe, S., Borgen, P.I. & Rimm, D.L. Vinculin is associated with the E-cadherin adhesion complex. J. Biol. Chem. 272, 32448–32453 (1997).
Hülsken, J., Birchmeier, W. & Behrens, J. E-cadherin and APC compete for the interaction with β-catenin and the cytoskeleton. J. Cell Biol. 127, 2061–2069 (1994).
Weiss, E.E., Kroemker, M., Rudiger, A.H., Jockusch, B.M. & Rudiger, M. Vinculin is part of the cadherin-catenin junctional complex: complex formation between α-catenin and vinculin. J. Cell Biol. 141, 755–764 (1998).
Tachibana, K. et al. Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J. Cell Biol. 150, 1161–1176 (2000).
Shapiro, L. et al. Structural basis of cell-cell adhesion by cadherins. Nature 374, 327–337 (1995).
Itoh, M., Nagafuchi, A., Moroi, S. & Tsukita, S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to α-catenin and actin filaments. J. Cell Biol. 138, 181–192 (1997).
Imamura, Y., Itoh, M., Maeno, Y., Tsukita, S. & Nagafuchi, A. Functional domains of α-catenin required for the strong state of cadherin-based cell adhesion. J. Cell Biol. 144, 1311–1322 (1999).
Pokutta, S. & Weis, W.I. Structure of the dimerization and β-catenin–binding region of α-catenin. Mol. Cell 5, 533–543 (2000).
Pokutta, S., Drees, F., Takai, Y., Nelson, W.J. & Weis, W.I. Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J. Biol. Chem. 277, 18868–18874 (2002).
Yang, J., Dokurno, P., Tonks, N.K. & Barford, D. Crystal structure of the M-fragment of α-catenin: implications for modulation of cell adhesion. EMBO J. 20, 3645–3656 (2001).
Choi, H.J. et al. αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc. Natl. Acad. Sci. USA 109, 8576–8581 (2012).
Peng, X., Maiers, J.L., Choudhury, D., Craig, S.W. & Demali, K.A. α-Catenin uses a novel mechanism to activate vinculin. J. Biol. Chem. 287, 7728–7737 (2012).
Jockusch, B.M. & Isenberg, G. Interaction of α-actinin and vinculin with actin: opposite effects on filament network formation. Proc. Natl. Acad. Sci. USA 78, 3005–3009 (1981).
Wilkins, J.A. & Lin, S. High-affinity interaction of vinculin with actin filaments in vitro. Cell 28, 83–90 (1982).
Johnson, R.P. & Craig, S.W. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature 373, 261–264 (1995).
Weekes, J., Barry, S.T. & Critchley, D.R. Acidic phospholipids inhibit the intramolecular association between the N- and C-terminal regions of vinculin, exposing actin-binding and protein kinase C phosphorylation sites. Biochem. J. 314, 827–832 (1996).
Steimle, P.A., Hoffert, J.D., Adey, N.B. & Craig, S.W. Polyphosphoinositides inhibit the interaction of vinculin with actin filaments. J. Biol. Chem. 274, 18414–18420 (1999).
Borgon, R.A., Vonrhein, C., Bricogne, G., Bois, P.R. & Izard, T. Crystal structure of human vinculin. Structure 12, 1189–1197 (2004).
Rangarajan, E.S., Lee, J.H., Yogesha, S.D. & Izard, T. A helix replacement mechanism directs metavinculin functions. PLoS ONE 5, e10679 (2010).
Xing, Y. et al. Crystal structure of a full-length β-catenin. Structure 16, 478–487 (2008).
Pappas, D.J. & Rimm, D.L. Direct interaction of the C-terminal domain of α-catenin and F-actin is necessary for stabilized cell-cell adhesion. Cell Commun. Adhes. 13, 151–170 (2006).
Johnson, R.P. & Craig, S.W. The carboxy-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochem. Biophys. Res. Commun. 210, 159–164 (1995).
Rimm, D.L., Koslov, E.R., Kebriaei, P., Cianci, C.D. & Morrow, J.S. α1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. USA 92, 8813–8817 (1995).
Knudsen, K.A., Soler, A.P., Johnson, K.R. & Wheelock, M.J. Interaction of α-actinin with the cadherin/catenin cell-cell adhesion complex via α-catenin. J. Cell Biol. 130, 67–77 (1995).
Pradhan, D., Lombardo, C.R., Roe, S., Rimm, D.L. & Morrow, J.S. α-Catenin binds directly to spectrin and facilitates spectrin-membrane assembly in vivo. J. Biol. Chem. 276, 4175–4181 (2001).
Kobielak, A. & Fuchs, E. α-Catenin: at the junction of intercellular adhesion and actin dynamics. Nat. Rev. Mol. Cell Biol. 5, 614–625 (2004).
Nagafuchi, A., Ishihara, S. & Tsukita, S. The roles of catenins in the cadherin-mediated cell adhesion: functional analysis of E-cadherin–α-catenin fusion molecules. J. Cell Biol. 127, 235–245 (1994).
Sako, Y., Nagafuchi, A., Tsukita, S., Takeichi, M. & Kusumi, A. Cytoplasmic regulation of the movement of E-cadherin on the free cell surface as studied by optical tweezers and single particle tracking: corralling and tethering by the membrane skeleton. J. Cell Biol. 140, 1227–1240 (1998).
Pacquelet, A. & Rorth, P. Regulatory mechanisms required for DE-cadherin function in cell migration and other types of adhesion. J. Cell Biol. 170, 803–812 (2005).
Chen, H., Cohen, D.M., Choudhury, D.M., Kioka, N. & Craig, S.W. Spatial distribution and functional significance of activated vinculin in living cells. J. Cell Biol. 169, 459–470 (2005).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011).
Kabsch, W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystalllogr. 26, 795–800 (1993).
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006).
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007).
Vagin, A. & Teplyakov, A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Bricogne, G. et al. BUSTER version 2.9 (Global Phasing Ltd., 2011).
Smart, O.S. et al. Refinement with local structure similarity restraints (LSSR) enables exploitation of information from related structures and facilitates use of NCS. Abstr. Annu. Meet. Am. Crystallogr. Assoc., Knoxville, TN, abstr. TP139 (2008).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Acknowledgements
We are indebted to our colleagues at Scripps Florida: J. Cleveland for discussions and critical review of the manuscript, Z. Wu and P. Bois for sequencing and P. Bois for fruitful discussions. We thank C. Vonrhein and G. Bricogne (Global Phasing Ltd.) for analyses and helpful discussions. We are grateful to the staff at the SER-CAT (BM22) and SSRL (11-1) for synchrotron support. T.I. is supported by grants from the US National Institute of General Medical Sciences from the US National Institutes of Health (GM071596 and GM094483) and by start-up funds provided to Scripps Florida from the State of Florida. This is publication no. 21863 from The Scripps Research Institute.
Author information
Authors and Affiliations
Contributions
Both authors contributed to the design and interpretation of all aspects of this work. E.S.R. performed all of the experiments. T.I. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Table 1 (PDF 39296 kb)
Rights and permissions
About this article
Cite this article
Rangarajan, E., Izard, T. Dimer asymmetry defines α-catenin interactions. Nat Struct Mol Biol 20, 188–193 (2013). https://doi.org/10.1038/nsmb.2479
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2479
This article is cited by
-
Interleukin-6 Downregulates the Expression of Vascular Endothelial-Cadherin and Increases Permeability in Renal Glomerular Endothelial Cells via the Trans-Signaling Pathway
Inflammation (2022)
-
α-Catenin levels determine direction of YAP/TAZ response to autophagy perturbation
Nature Communications (2021)
-
Binding partner- and force-promoted changes in αE-catenin conformation probed by native cysteine labeling
Scientific Reports (2019)
-
Real-time TIRF observation of vinculin recruitment to stretched α-catenin by AFM
Scientific Reports (2018)
-
Force-dependent allostery of the α-catenin actin-binding domain controls adherens junction dynamics and functions
Nature Communications (2018)