Abstract
Signal transducer and activator of transcription 3(STAT3) is an emerging target for cancer therapy. In this study, we identify Toosendanin (TSN) is an effective inhibitor of STAT3, leading to the impediment of various oncogenic processes in osteosarcoma. TSN selectively inactivates phospho-STAT3 (Tyr-705); subsequent molecular docking and in vitro SPR analysis uncover TSN directly binds to the SH2 domain of STAT3. Consequently, TSN blocks STAT3 dimerization and impairs the complex formation of STAT3 and epidermal growth factor receptor (EGFR). In an animal tumor model study, TSN is well tolerated, inhibits osteosarcoma growth and metastasis. In another osteosarcoma patient-derived xenografts (PDX) model, we find TSN triggers strong inhibitory effects on patient-derived tumors. Further studies show that TSN also displays activity against other solid tumors. Our preclinical work therefore supports that TSN acts as a novel inhibitor of STAT3 that blocks tumorigenesis in ostoesarcoma.
Similar content being viewed by others
Introduction
Constitutive activation of the signal STAT3-meditated signal pathway plays pivotal roles in tumor cell growth, survival, apoptosis, angiogenesis and metastasis.1, 2 Mounting evidence demonstrates that constitutively activated STAT3 contributes to tumor development and progression in the majority of cancers, including breast, prostate, ovary, lung, gastric, melanoma and blood.3, 4 Constitutively activated STAT3 correlates with a more malignant tumor phenotype and is related with decreased survival in some cancers.5 Interestingly, in contrast to their cancerous counterparts, non-cancerous cells do not employ constitutively activated STAT3 to maintain their growth, and many studies have supported that they are not sensitive to loss of STAT3 function or STAT3 inhibitors.6 Therefore, STAT3 is deemed as an attractive target for antitumor drug development.
In support of these backgrounds, several approaches have been proposed to suppress constitutive activation of STAT3 and varieties of STAT3 inhibitors have been designed and discovered. Inhibitors of STAT3 can be split into two categories, which are: direct and indirect. Indirect inhibitors interfere with its ligands such as cytokines (IL-6, IL10 and so on) and growth factor receptors (VEGFR, IGFR, EGFR and so on), or upstream kinases (JAKs and Src) that phosphorylate STAT3.7, 8 Inhibitors directly interact with the STAT3 protein can be distinguished based on the distinct binding domain, for example, the NTD, DBD or SH2 domains of STAT3.9 The SH2 domain of STAT3 is involved in upstream receptor kinases recognition and subsequent STAT3 dimerization.10 Induced by tyrosine phosphorylation, STAT3 dimerization is a prerequisite for DNA binding. Given its critical role in STAT3 activation and function, the SH2 domain has been considered as the most attractive targetable site of STAT3. SH2-targeting compounds take the largest proportion of direct STAT3 inhibitor such as OPB-31121, an oral STAT3 inhibitor undergoing phase I/II clinical trials in hepatocellular carcinoma.11
Osteosarcoma represents the most frequently diagnosed malignancy in children and adolescents, and arises from primitive bone-forming mesenchymal cells.12 Despite significant advances in surgery and multiagent chemotherapy, nearly 30% of patients still die from osteosarcoma.13 Therefore, it is necessary to develop novel therapeutic approaches for osteosarcoma treatment. Compelling evidence from previous studies has demonstrated the key role o STAT3 in osteosarcoma development and STAT3 might become an attractive molecular target for drug discovery of human osteosarcoma.14, 15
Agents derived from natural sources have gained much attention because of their safety, efficacy and immediate availability, and they are the best sources of drugs and drug leads for novel drug discovery.16 Some natural products and derivatives have been found to possess inhibitory function on STAT3 activation such as curcumin,17 resveratrol18 and others. However, the specific molecular basis underlying the suppressive effects of these agents on STAT3 remains unveiled. Here we found that Toosendanin (TSN), a triterpenoid saponin from the bark of the trees Melia toosendan and M azeduvach (Meliaceae),19 directly binds to the STAT3-SH2 domain, thus exerting significant anti-STAT3 signaling effect at nanomolar concentration. In addition, we demonstrate the efficacy of TSN in osteosarcoma using both in vitro and in vivo models. These data provide the proof-of-concept for evaluating STAT3 inhibitors as anti-osteosarcoma agents.
Results
TSN is a potent inhibitor of STAT3 tyrosine phosphorylation and downregulates STAT3 downstream target genes expression
STAT3 is a transcription factor that regulates genes involved in cell growth, metastasis and angiogenesis, and it has recently emerged as a promising target for cancer therapy. A STAT3 luciferase reporter assay was used to identify novel STAT3 signaling inhibitors. Using a preliminary screening of our internal chemical library, we identified the natural product toosendanin (TSN, MW: 574.62) as a putative hit for blocking of the STAT3 signaling. TSN showed potent STAT3 inhibitory activity in osteosarcoma cell lines (Figures 1a and b). To further confirm the STAT3 inhibitory effect, we detected the constitutive activation of STAT3 in osteosarcoma cells using specific antibodies against phospho-STAT3 (Tyr-705) and phospho-STAT3 (Ser727). We found TSN significantly blocked phospho-STAT3 (Tyr-705) activation at 100 nM; however, in the same condition, phospho-STAT3 (Ser727) and STAT3 protein expression remained unchanged (Figure 1c). To regulate its target genes expression, STAT3 must translocate from the cytosol to the nucleus. 143B cells were exposed to TSN for 24 h, and then stimulated with IL-6. As shown in Figure 1d, STAT3 nuclear translocation was largely inhibited when exposed to TSN. In addition, an electrophoretic mobility shift assay (EMSA) demonstrated that treatment with TSN led to a dose-dependent inhibition of STAT3 DNA-binding activity in 143B cells (Figure 1e). Protein tyrosine phosphatases (PTPase) are reported to involve in the inactivation of STAT3. We next set forth to investigate whether TSN could block STAT3 activation through a certain PTPase.20 All three known PTPases (SHP1, SHP2 and PTEN) mediating STAT3 activity remain constant after TSN treatment, indicating PTPases do not contribute to the STAT3 inhibition by TSN (Supplementary Figure S1a). To further establish whether TSN exclusively inactivated STAT3, STAT1, STAT2, STAT5, MAPK and mTOR signaling pathways were evaluated. In 143B cells, the status of STAT1, STAT2, STAT5, p38, ERK, mTOR and p70S6 remained unchanged after addition of TSN (Supplementary Figure S1b and c). The aforementioned results showed that TSN perturbed phosphorylation, nuclear translocation and DNA-binding activity of STAT3. We next determined whether TSN affected STAT3 transcriptional activity. The translocation of STAT3 dimers to the nucleus results in their specific binding to target gene promoters, thus rewiring gene expression. STAT3 participates in oncogenesis through mediating its target gene, including Bcl-2, Mcl-1, survivin, VEGF and MMPs.21 Previous studies also indicated that activation of STAT3 plays an essential role in SOCS3 expression.22 The analysis of the target gene products of STAT3 revealed that TSN significantly reduced its target gene expression at both the mRNA and protein level (Figures 1f and g). ChIP assays further revealed that TSN disturbed interaction between STAT3 and its target genes (Figure 1h). In conclusion, our results demonstrated TSN specifically blocked STAT3 tyrosine phosphorylation and repressed the expression of the STAT3 downstream targets in osteosarcoma.
TSN selectively inhibits STAT3 activity in osteosarcoma. (a) Osteosarcoma cells were transfected with luciferase reporter gene plasmid and treated with TSN for 24 h. The results were normalized to the Renilla luciferase activity. The bars indicate the mean±s.d. Statistically significant differences (t-test), *P<0.05; **P<0.01; ***P<0.001. (b) The chemical structure of TSN. (c) After treatment with TSN for 24 h, cell extracts were prepared and applied to immunoblotting with phospho-STAT3 (Tyr-705) or phospho-STAT3 (Ser727). Actin was used as a loading control. (d) Left panel, 143B cells were cultured on gelatin-coated coverslips and treated with TSN for 24 h followed by stimulating with IL-6 (20 ng/μl) for 30 min. Anti-STAT3 antibody (green) was used to locate endogenous STAT3. Cell nuclei were stained with 4′, 6-diamidino-2-phenylindole (DAPI). The photographs were acquired by a confocal microscopy. Scale bar, 20 μm. Right panel, l43B cells were treated with TSN for 24 h, followed by stimulating with IL-6 (20 ng/μl) for 30 min, and the cytoplasmic and nuclear extractions were subjected to immunoblotting to detect the distribution of STAT3. (e) 143B cells were pretreated with TSN. An EMSA assay was performed to analyze STAT3 DNA-binding activity. (f) Cells were treated with TSN for 24 h. STAT3 target genes expression were analyzed by RT-PCR. (g) Cells were treated with TSN for 24 h. Cells were then lysed and applied to immunoblotting with indicated antibodies. Actin was used as a loading control. (h) 143B cells were incubated with TSN and analyzed by a quantitative ChIP assay with anti-STAT3 antibody. The bars indicate the mean±s.d. Statistically significant differences (t-test), *P<0.05; ***P<0.001.
TSN directly binds the SH2 domain of STAT3
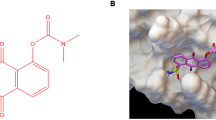
To explore whether TSN interacted with STAT3, a molecular docking model of TSN with X-ray crystal structure of STAT3 (PDB Id: 1BG1) were established by the Autodock 4.2 and UCSF Chimera 1.7. Our computational modeling results showed that TSN was docked into the SH2 domain of STAT3 (Figure 2a and Supplementary Figure S2a). To further verify whether TSN interacts directly with STAT3-SH2 domain, we purified GST-tagged STAT3-SH2 domain from Escherichia coli (Figure 2b). The binding between TSN and STAT3-SH2 domain was evaluated by surface plasmon resonance (SPR) assay using a biacore platform. As shown in Figure 2c, the RU values evaluating the TSN binding to the immobilized STAT3-SH2 protein demonstrated a dose-dependent manner and the determined equilibrium dissociation constant (KD) is about 240 nM. STAT3 monomers form activated dimmers through reciprocal phosphotyrosine SH2 interaction between monomers. A GST pull-down assay demonstrated the interplay between purified STAT3-SH2 and STAT3 in cell extracts was significantly inhibited by TSN (Figure 2d). To further confirm TSN suppresses STAT3–STAT3 dimerization, we constructed HA-tagged and Flag-tagged STAT3 vector, respectively. The vectors were transiently transfected into 293 T cells. Figure 2e indicated that Flag-STAT3 co-immunoprecipitated with HA-STAT3 in 293 T cells and TSN disrupted the interaction in a concentration-dependent manner. We also found that TSN significantly blocked STAT3:STAT1 heterodimers formation (Supplementary Figure S3a). These results suggested that TSN binds to the STAT3-SH2 domain, further blocking STAT3:STAT3 homodimerization and STAT3:STAT1 heterodimerization. STAT3-SH2 domain can also bind to some receptor tyrosine kinases such as EGFR, which in turn leads to its activation.23 Using an in vitro pull-down assay, we found purified STAT3-SH2 interplays with EGFR and TSN treatment inhibited the complex formation (Figure 2f). We next evaluated if TSN could lead to the endogenous STAT3-EGFR complex dissociation. We found that stimulation osteosarcoma cells with EGF increased the capability of EGFR to bind to STAT3 and treatment with TSN inhibited these interactions (Figure 2g). These results suggested that TSN directly targets STAT3-SH2 domain.
TSN directly binds the SH2 domain of STAT3. (a) Molecular docking model carried out by the autodock 4.2 and UCSF Chimera 1.7 revealed that TSN binds to the SH2 domain of STAT3. (b) SDS PAGE analysis of purified GST-tagged STAT3-SH2 protein. Protein band (about 42 KD) was visualized by coomassie blue staining. (c) SPR analysis of the binding between TSN and STAT3-SH2 domain. The purified STAT3-SH2 protein was immobilized on an activated CM5 sensor chip. TSN was then flowed across the chip. (d) TSN blocked SH2–SH2 interaction. Purified STAT3-SH2 protein was mixed with different concentration TSN for 2 h, followed by incubation with 143B cell lysates. A GST pull-down analyses were performed. (e) TSN inhibited STAT3–STAT3 dimerization. 293 T cells transfected with FLAG and HA-tagged STAT3 vectors were treated with TSN at the indicated concentration, subjected to immunoprecipitation, and immunoblotted with FLAG or HA antibodies. (f) Purified STAT3-SH2 protein was mixed with different concentration TSN for 2 h, followed by incubation with 143B cell lysates. A GST pull-down assay was performed. (g) Cells (143B or HOS) were treated with different concentrations of TSN and stimulated with or without EGF, followed by co-immunoprecipitation assays.
TSN suppresses osteosarcoma cells growth and angiogenesis
Our experiments up to this point clearly indicated that TSN is an effective STAT3 inhibitor. To assess the antitumor function of TSN in vitro, we first evaluated its inhibitory effects on cell proliferation. TSN suppressed osteosarcoma cell viability in a robust and concentration-dependent manner (Figure 3a). We also found TSN showed no obvious cytotoxicity against hFOB1.19 normal osteoblast cells (Figure 3a). This result suggested that the use of TSN in cancer treatment may be safe and selectively cytotoxic to osteosarcoma cells. Western blot assays indicted the phospho-STAT3 (Tyr-705) level was reduced in TSN-treated osteosarcoma cell lines but remained unchanged in normal osteoblast cells (hFOB1.19) (Figure 1c and Supplementary Figure S4a). To further investigate if the STAT3 signaling is involved in TSN-induced growth arrest, we transfected STAT3 vector to rescue TSN-induced STAT3 inhibition. STAT3 overexpression significantly impaired TSN-induced growth arrest and STAT3 target gene inhibition (Figure 3b). We also studied the effect of TSN on colony formation, since it mimics tumor physiology and growth in vivo. Osteosarcoma cell colony formation was greatly impeded by TSN exposure (Figure 3c). In addition, Annexin V/7AAD assays showed TSN (50 nM) significantly induced cell apoptosis. In accordance with these data, cleaved PARP was accumulated, whereas PARP was reduced under TSN treatment (Figure 3d). Pathological angiogenesis is a hallmark of cancer and is closely associated with tumor development.24 To evaluate the anti-angiogenensis efficacy of TSN in osteosarcoma, a tubular formation assay was performed. TSN exerted little effect on HUVECs viability (Supplementary Figure S5a), and capillary-like structure formation was not affected by TSN in culture medium at the same concentration (Supplementary Figure S5b). In contrast, when exposure to conditional medium from TSN-treated 143B cells, HUVECs could not form integrated tubular structure (Figure 3e). Collectively, these data indicated that TSN suppresses osteosarcoma cells growth and angiogenesis.
TSN suppresses osteosarcoma cells growth and angiogenesis. (a) Osteosarcoma cell lines or normal osteoblast cell hFOB1.19 was exposed to indicated concentrations of TSN for 48 h and a MTS assay was performed. The bars indicate the mean±s.d. Statistically significant differences (Student’s t-test), **P<0.01; ***P<0.001. (b) 143B cells were transfected with STAT3 vector, and then a MTS, western blot and qPCR assay were performed. (c) Osteosarcoma cells were seeded into six-well plates and treated with or without 25 nM TSN for a week. Colonies were then fixed, stained and photographed. Colony numbers were counted manually. The bars indicate the mean±s.d. Statistically significant differences (Student’s t-test), **P<0.01; ***P<0.001. (d) Cells were treated with indicted concentrations of TSN for 48 h. Cells were stained with Annexin V and 7AAD, and analyzed by flow cytometry. The protein level of PARP and cleaved PARP were detected via western blot assay. (e) Tube formation assay. HUVECs (2 × 104) were seeded on Matrigel with conditional medium. After incubation for 8 h, photomicrographs were acquired with an inverted microscope (OLYMPUS). Scale bar, 40 μm. The bars indicate the mean±s.d. Statistically significant differences (Student’s t-test), **P<0.01; ***P<0.001.
TSN inhibits osteosarcoma cells invasion and reserves EMT phenotype
Metastasis represents the major conundrum in osteosarcoma therapy. During metastasis, tumor cells must invade surrounding tissues and intravasate through the endothelium to blood or lymphatic circulation.25 We performed invasion assays using two highly metastatic human osteosarcoma cell lines 143B and MG63.2. We found that TSN suppressed tumor cell invasion in a concentration-dependent manner (Figure 4a). The degradation of basement membrane and extracellular matrix (ECM) is the prerequisite for the invasion and intravasation processes.26 We next conducted a fluorescent-gelatin degradation assay to test whether TSN inhibits osteosarcoam cells to degrade ECM. The result showed that 143B cells potently degraded ECM in 12 h (control group), while TSN-treated group failed to do so (Figure 4b). Three-dimensional culture systems provide structural and functional aspects of cancer progression. In our three-dimensional culture model, TSN significantly inhibited tumor cell invasion through the surrounding Matrigel (Figure 4c). Epithelial–mesenchymal transition (EMT) is the initial and key step toward cancer metastasis.27 During EMT, tumor cell phenotype changes with a series of proteins, including epithelial and mesenchymal markers. We found TSN significantly downregulated the expression of N-cadherin, fibronectin and vimentin while upregulated E-cadherin (Figures 4d and e). Together, these results indicated that TSN shows metastasis inhibitory effects in vitro.
TSN inhibits osteosarcoma cells invasion and reserves EMT phenotype. (a) 143B and MG63.2 cells were seeded into the upper chamber of the transwell inserts. After 12 h incubation, images were obtained, Scale bar, 40 μm. Cell invasion was quantified manually. The bars indicate the mean±s.d. Statistically significant differences (Student’s t-test), *P<0.05; **P<0.01; ***P<0.001. (b) 143B cells were seeded on FITC-conjugated gelatin (green) for 12 h. Phalloidin (red) was used to indicate F-actin and nuclei was stained with DAPI (blue). Gelatin degradation areas were the black areas underneath the cells, Scale bar, 10 μm. The bars indicate the mean±s.d. Statistically significant differences (Student’s t-test), ***P<0.001. (c) 3D culture assay. 143B cells were plated onto a layer of Matrigel. DMEM plus 10% Matrigel as well as indicated concentrations of TSN was then added. The on-top mixture was changed every other day. Four days later, cells were photographed, Scale bar, 40 μm. (d) After treatment with TSN for 72 h, a western blot assay was performed. (e) After treatment with TSN for 72 h, the level of N-cadherin (green) and E-cadherin (green) were detected by IF staining. Nuclei were stained with DAPI (blue). Scale bar, 10 μm.
TSN inhibits tumor growth, metastasis and prolongs the survival of mice in osteosarcoma xenografts
To determine whether TSN inhibits osteosarcoma growth and metastasis in vivo, we established tumor xenograft in the medullary cavity of tibia of each mouse with 143B cells. When tumors were palpable, mice were randomized to receive either control (DMSO) or TSN by every other daily injection. As shown in Figure 5a, TSN administered at 1 mg/kg or 2 mg/kg significantly inhibited osteosarcoma growth, as tumor weight was markedly relived in TSN treatment groups compared with control group. Notably, the number of lung metastasis nodules was significantly reduced in mice that received TSN (Figures 5b and c). In order to carry out mechanism research, tumor tissues were either prepared for western blot or immunohistochemistry (IHC) analysis. We found TSN inhibited phospho-STAT3 (Tyr-705) and target genes expression in low- and high-dose groups (Figure 5d). IHC analysis also showed that TSN significantly reduced phospho-STAT3 level in vivo (Figure 5e, upper panel). Moreover, we also found the reduction of tumor vascular density in TSN treatment group (Figure 5e, lower panel). To evaluate whether the metastasis suppression effect of TSN could arouse a survival benefit, the survival rate of tumor-bearing mice was analyzed. Our analysis indicated that TSN remarkably prolonged overall survival of mice. On day 40, all the mice had died in control group (n=8), whereas only two mouse died in high-dose TSN treatment group (Figure 5f). To our surprise, there were still six mice alive in high-dose TSN (2 mg/kg) group on day 50. Taken together, these data demonstrated that TSN is detrimental to the growth and metastasis of osteosarcoma in the preclinical osteosarcoma xenograft model.
TSN inhibits tumor growth, metastasis and prolongs the survival of mice in an orthotopic osteosarcoma mouse model. 143B tumor cells (1 × 106) were injected into the medullary cavity of tibia of each mouse. Seven days later, the mice were randomly allocated into three groups (n=6 per group) and received i.p. injection of TSN. After 24 days, all mice were executed. (a) The weight of the posterior limb with tumors was measured. Statistically significant differences (Student’s t-test), **P<0.01; ***P<0.001. (b) Lung colonization was visualized by a dissecting microscope and lung metastasis nodules were counted manually. Statistically significant differences (Student’s t-test), **P<0.01; ***P<0.001. Scale bar, 200 μm. (c) Representative images of lung tissue in control and treatment groups (upper panel). 4 μm sections of lungs were subjected to H&E staining (lower panel). (d) The tumor tissues were extracted in RIPA lysis buffer, a western blot assay was performed. (e) Representative pictures of p-STAT3 and CD31 immunostaining in tumors. Scale bar, 20 μm. (f) Overall survival rate according to treatment in the orthotopic osteosarcoma mouse model was shown. Time scale relates to days after treatment. TSN vs Control (1 mg/kg), P=0.0002; TSN vs Control (2 mg/kg), P<0.0001.
Antitumor activity of TSN in a patient-derived osteosarcoma xenograft animal model
Patien-derived xenograft (PDX) animal models may be superior to standard cell-line xenograft models of cancer because they maintain more similarities to the parental tumors.28 A PDX model is an attractive option for testing of novel compounds.29 Seven osteosarcoma specimens along with the corresponding adjacent normal tissue were obtained at initial surgery from primarily diagnosed and the information of patients was listed in the supplementary table 2. To explore the potential role of STAT3 in osteosarcoma tumorigenesis, we first examined the expression of phospho-STAT3 (Tyr-705) and STAT3 in these clinical specimens. We found phospho-STAT3 (Tyr-705) and STAT3 were hyperactivated in tumor tissues compared to normal ones (Figure 6a). These data indicated that increased STAT3 activation is a common event in osteosarcoma. To establish the PDX animal model, we devised a procedure as shown in Figure 6b (Figure 6b, left panel). In these samples, 3 passagable xenograft models (2#, 4# and 7#) were successfully established. We selected 4# patient-derived xenograft animal model for our following experiments. We also detected the expression of phospho-STAT3 (Tyr-705) and STAT3 at the protein level in different passages of 4# patient-derived xenograft. The expression level of phospho-STAT3 (Tyr-705) showed no obvious differences in these passages (F0, F1, F2 and F3) (Figure 6b, right panel). This result implied that the highly active characteristics of STAT3 still remain when continuous passage in mice. The third-generation mice were randomly divided into three groups. We detected an obvious decrease in tumor growth among TSN-treated and control groups (Figure 6c). Tumors treated with TSN had an average volume of 608.95 mm3 (1 mg/kg per 2 days) and 372.17 mm3 (2 mg/kg per 2 days). However, the average volume of control group was 1394.49 mm3 (Figure 6d). Tumor weight was significantly reduced in TSN-treated groups (Figure 6e). Notably, the expression of phospho-STAT3 (Tyr-705) and STAT3 targets (Survivin, Bcl-2 and VEGF) were downregulated after TSN treatment, suggesting that STAT3 is still a major target of TSN in vivo (Figure 6f). Immunohistochemical analyses of tumor tissues from animal model revealed low phospho-STAT3 (Tyr-705) and tumor vascular density level after TSN treatment (Figure 6g). Taken together, these data indicated that the antitumor effects of TSN are due to inhibition of STAT3 in PDX animal model and supported TSN may display potential therapeutic effects in clinical trials.
Antitumor activity of TSN in a patient-derived osteosarcoma xenograft animal model. (a) Seven human osteosarcoma tumors and their paired adjacent normal muscular tissues were tested. Expression of p-STAT3 (Tyr-705) and STAT3 in the tissue extracts was determined using specific antibodies. (b) Establishment of a PDX model (left panel). Expression of p-STAT3 (Tyr-705) and STAT3 was determined in different passages of PDX. (c) Representative images of tumor tissue in control and treatment groups of PDX model (n=8). (d) The PDX tumor volume was shown (n=8, ***P<0.001). (e) The result of tumor weight in control group and TSN-treated group was shown (n=8, ***P<0.001). (f) The tumor tissues were extracted, and a western blot assay was executed. (g) Representative pictures of p-STAT3 and CD31 immunostaining in tumors. Scale bar, 20 μm.
TSN suppresses the growth of other tumor cell lines
Considering that STAT3 is essential for tumor development and progression in several cancer types, we wondered whether TSN could repress other tumor cell growth. As showed in Figure 7a, TSN treatment led to the growth arrest of other tumor cell lines. We also treated these cell lines with TSN and evaluated its inhibitory effects on STAT3 signaling (Figure 7b). Phospho-STAT3 (Tyr-705) activation was largely blocked by TSN in these non-osteosarcoma cancer cells.
TSN inhibits the growth of other tumor cell lines. (a) Various cancer cell lines were treated with TSN (100 nM and 200 nM). After 48 h, viable cells were measured using a MTS assay. The bars indicate the mean±s.d. Statistically significant differences (Student’s t-test), **P<0.01; ***P<0.001. (b) Tumor cells were exposed to TSN (100 nM and 200 nM) for 24 h. Samples were immunobloted using the indicated antibodies.
The potential toxicity of TSN on mice
We examined the potential toxicity of TSN (2 mg/kg per 2 days). Male BALB/c mice received intraperitoneal injection of TSN for 28 days (n=5). Body weight was measured once a week. When the experiment terminated, the major organs were embedded for H&E staining. No significant changes in mice body was found after treatment (Supplementary Figure S6a). In addition, the result of H&E staining demonstrated no significant histological changes between control and TSN-treated group (Supplementary Figure S6b). Together, these results implied that TSN shows few side effects on the mouse body at our therapeutic concentration.
Discussion
The discovery of small molecular compounds for treating osteosarcoma is important to improve patient outcome. Here we identify a novel STAT3 inhibitor, Toosendanin, in our internal traditional Chinese medicine compounds library using a STAT3 luciferase reporter assay. We combined in silico and in vitro experiments to prove TSN directly binds to STAT3-SH2 domain of STAT3, and we also provided the first demonstration that TSN exhibited potent antiumor activity in osteosarcoma cell lines and mouse xenograft models via inhibiting STAT3 signaling.
Src homology 2 (SH2) domains is highly conserved regions common to STAT3, and involved in receptor recognition as well as in their dimerization.30 STAT3 dimerization is induced by its tyrosine phosphorylation and is obligatory for nuclear translocation and subsequent DNA binding. Molecular modeling studies give solid support to the fact that TSN binds to STAT3-SH2 domain. In addition, our molecular modeling demonstrated that TSN makes directly interact with Arg-609 and Lys-591 residues (Figure 2a and Supplementary Figure S2a), which are critical amino acids that are known to bind phospho-Tyr-705 in the SH2 domain of STAT3.31 Indeed, surface plasmon resonance (SPR) assay further supported our docking model. We also found TSN disrupted STAT3:STAT3 homodimerization and STAT3:STAT1 heterodimerization as shown by co-immunoprecipitation analysis (Figures 2d and e and Supplementary Figure S3a). Furthermore, previous studies reported that STAT3 interact with EGFR through its SH2 domain, and the bound EGFR reciprocally phosphorylates STAT3 and induces its dimerization. Our data indicated that TSN significantly inhibited STAT3-EGFR complex formation in our system (Figures 2f and g).We speculate that TSN may also disturb the binding of STAT to other nonreceptor tyrosine kinases, such as Janus-activated kinase (JAK) 2 and Src, which are considered crucial inducers for STAT3 dimerization. Therefore, we hold that TSN is a STAT3 inhibitor via binding to STAT3-SH2 domain, further causing the dysfunction of STAT3 and its downstream cascade. To our surprise, TSN has no obvious effect on the phosphorylation status of Ser727 site of STAT3. STAT3 transcriptional activation could be regulated by phosphorylation at Ser727 through the MAPK or mTOR pathways. We also found that TSN have little effect on MAPK and mTOR signaling. These phenomena may explain why TSN has no effect on the phosphorylation of STAT3 at Ser727 site. These data implies TSN is a novel STAT3 inhibitor and has a specific inhibitory action against phospho-Tyr-705 of STAT3.
TSN inhibited tumor cell growth as well as suppressed the expression of genes that are known to drive uncontrolled cellular growth of cancer, such as Bcl-2, Mcl-1 and survivin. In our study, TSN showed little effect on other signaling pathways such as STAT1, STAT2, STAT5, MAPK and mTOR, suggesting that TSN induces cell proliferation arrest through inhibition of STAT3. Previous studies indicated STAT3 activation is rapid and transient in non-cancerous cells, but constitutively activated in the majority of cancers.6 This implies constitutively activated STAT3 is essential for cancer development at difference levels. In line with this, our work uncovered TSN significantly inhibits osteosarcoma cell growth at a nanomolar concentration and has little cytotoxicity in two normal cell lines hFOB1.19 osteoblast cells and HUVECs. Our results also demonstrated TSN selectivity inhibits phosphorylation of STAT3 in osteosarcoam cells. Vascular endothelial growth factor (VEGF) is a critical inductor of pathological angiogenesis associated with tumors.32 Previous studies reported VEGF expression positively correlated with STAT3 activity in various human cancers and VEGF is proved to be a target gene of STAT3.33 In our study, we found TSN significantly downregulated VEGF at the mRNA and protein level (Figures 1f and g). ChIP assays indicated that STAT3 directly regulated VEGF expression and TSN significantly inhibited STAT3 binding to the VEGF promoter region (Figure 1h). Using conditional medium from osteosarcoma cells, we found that TSN suppressed tubular-like structure formation in a dose-dependent manner (Figure 3e); however, TSN showed no effect on capillary structure formation in cell culture medium at the same concentration inducer (Supplementary Figure S5b). We also observed the reduction of tumor vascular density (CD31) in TSN treatment mice in vivo. Therefore, we hold that, the reason that TSN inhibits tumor angiogenensis is it could dysregulate the function of tumor cellular secreting growth factor, such as VEGF. These results support that TSN suppresses tumor growth by selective inhibition of STAT3 signaling pathway.
Tumor metastasis is a complex cascade during which cancer cells disseminate from primary tumors and successfully reside in secondary tumors in target organs.34 One of the key steps promoting tumor metastasis is ECM degradation. Matrix metalloproteinase-2 (MMP2) is a known target of STAT3 and it acts as an invasion, angiogenesis, and metastasis-related protein. We showed that TSN significantly inhibited MMP2 expression in gene and protein level (Figures 1f and g). In addition, degradation of matrix was suppressed after treating with TSN (Figure 4b). EMT is a prerequisite for cancer metastasis. Highly metastatic propensity of osteosarcoma may be partly due to its mesenchymal origin. Previous studies reported STAT3 plays essential roles in tumor EMT process and silence of STAT3 significantly increased E-cadherin and decreased N-cadherin expression in CRC cells.35 In our studies, we found TSN also significantly regulated EMT-related protein level in osteosarcoma (Figures 4d and e). In a cell line-derived xenograft animal model, TSN significantly inhibited osteosarcoma lung metastasis and prolonged the survival of mice. These results demonstrate that TSN serves as a novel STAT3 inhibitor that inhibits MMPs expression and reversed osteosarcoma cells EMT, thereby suppressing osteosarcoma metastasis.
One of the important findings in this study is that TSN displayed satisfactory therapeutic effect in an osteosarcoma PDX animal model. PDX models maximally retain the principal histological and genetic characteristics of patient tumor sample and are being used for preclinical drug research and personalized medicine strategies. We found phospho-STAT3 (Tyr-705) was continuously activated in tumor compared to normal tissue (Figure 6a) More importantly, osteosarcoma PDX animal model maintain the hyperactivation of phospho-STAT3 (Tyr-705) through serial passaging (F0, F1, F2 and F3) in mice (Figure 6b, right panel). Surprisingly, mice treated with TSN showed a robust inhibition of tumor growth. These results showed that TSN may provide significant clinical benefits in the treatment of osteosarcoma and our study provides solid evidence supporting that TSN possesses potential therapeutic effects in future clinical trials.
Our studies demonstrated that TSN exhibits the inhibitory potential for osteosarcoma growth and metastasis by directly binding to STAT3-SH2 domain, thereby inhibiting STAT3 activation. Yet, it is still plausible that TSN may have other targets involved in tumor progression. In addition, previous studies reported that high dose of TSN caused most serious liver injury (80 mg/kg) in mice.36 In our preliminary toxicity experiment, we found TSN has few side effects on the mouse body at our therapeutic concentration (2 mg/kg). Further investigations are needed to comprehensively explore the pharmacokinetics and pharmacodynamics of TSN, which can further guide us to use TSN for osteosarcoma therapy. Also, future studies may expand its role in combination with chemotherapy for osteosarcoma.
Materials and methods
Materials
Purified Toosendanin (MF: C30H38O11, MW: 574.62, purity >98%) was purchased from Shanghai Yuan Ye Biotechnology Co. Ltd (Shanghai, China). All the cell culture reagents were purchased from Invitrogen Life Technologies (Carlsbad, CA, USA). Dimethyl sulfoxide (DMSO) was obtained from Sigma-Aldrich (St Louis, MO, USA). Matrigel was purchased from BD Bioscience (Pasadena, CA, USA). p-STAT3 (Y705)(#9145), p-STAT3 (S705)(#94994), STAT3 (#9139), Survivin (#2808 S), Bcl-2 (#15071), SOCS3 (#2923), PTEN (#9188), PARP (#9532), Cleaved PARP (#5625), N-cadherin (#14215), Zeb1 (#3396)and E-cadherin (#14472) antibodies were purchased from Cell Signaling Technology Inc. (Danvers, MA, USA). Antibodies of SHP-1 (ab32559), SHP-2 (ab32083) and fibronectin (ab2413) were purchased from Abcam (Hong Kong, China). Antibody of actin (A1978) was purchased from Sigma-Aldrich (Sigma-Aldrich, Inc., Shanghai, China).
Cell culture
Cell lines 143B (human osteosarcoma cell), HOS (human osteosarcoma cell), MG63 (human osteosarcoma cell), HCT116 (human colon cancer cell), SGC7901 (human gastric carcinoma cell), SKOV3 (human ovarian cancer cell), MDA-MB231 (human breast cancer cell), DU145 (human prostate cancer cell), HeLa (human cervical cancer cell) and SW1990 (human pancreatic cancer cell) were purchased from ATCC (Manassas, VA, USA). MG63.2 cell line was established by serially passaging the parental MG63 cells.37 The murine LM8 osteosarcoma cell line and its parental cell line Dunn with low metastatic potential were originally established by Prof. Tatsuya Asai (Department of Orthopaedic Surgery, Osaka Medical Center for Cancer and Cardiovascular, Diseases, Osaka, Japan). All cell lines were maintained in DMEM supplemented with 10% FBS plus 1% penicillin/streptomycin. HUVECs (ScienCell Research Laboratories, San Diego, CA, USA) were cultured in complete ECM (Sciencell) supplemented with 5% FBS. hFOB1.19 cells were purchased from and cultured in DMEM/F12(1:1) with 10% FBS and 300 ng/mLG418. Cells were maintained at 37 °C under a humidified 5% CO2 incubator. Mycoplasma contamination was monitored periodically.
STAT3 luciferase reporter assay
The STAT3 luciferase reporter plasmid (pGMSTAT3-Luc) was used to detect STAT3 activation and obtained from Shanghai Yi Sheng Biotechnology Co. Ltd. and the assay was carried out as previously described.3 Briefly, cells were co-transfected with pGMSTAT3-Luc and pRL-SV40 (a plasmid encoding Renilla luciferase) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 24 h, cells were treated with test compounds for 24 h. Luciferase activity was assessed by the dual-luciferase reporter assay system (Promega, Madison, WI, USA) using a luminometer (Thermo Scientific, Waltham, MA, USA).
Western blot assay
Western blot analysis was performed as previously described.38 Proteins were extracted in RIPA cell lysis buffer containing protease and phosphatase inhibitors. Immunoblotting was performed with indicated antibodies.
Immunofluorescent staining
Cells were cultured and treated on glass coverslip, and fixed with 4% paraformaldehyde followed by 5 min of permeabilization with 0.1%Triton X-100. After incubation with indicted antibodies overnight at 4 °C, cells were incubated with corresponding secondary antibodies for 1 h at room temperature, and then stained with DAPI and visualized with confocal microscope (Leica).
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using Odyssey Infrared STAT3 EMSA Kit (LI-COR Biosciences, Lincoln, NE, USA) according to the manufacturer’s instructions.39 STAT3 IRDye 700 infrared dye-labeled oligonucleotides were: 5′-GATCCTTCTGGGAATTCCTAGATC-3′ and 3′-CTAGGAAGACCCTTAAGGATCTAG-5′ (boldface indicates STAT3-binding sites).
RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). RNA samples (1 μg) were subjected to RT-PCR using the TaKaRa RT-PCR kit (Takara, Shiga, Japan). Then PCR products were detected by electrophoresis in 1% agarose gels and were visualized using GoldView. The primers used are listed in the Supplementary Table 1.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as previously described.3 In brief, after treatment, 143B cells were treated with 1% formaldehyde for 10 min to crosslink chromatin and protein. The chromatin-protein samples were immunoprecipitated with STAT3 antibody. The immuoprecipitates were then incubated with protein A/G agarose beads. After several washes, the protein–DNA complex was reversed. DNA was purified using phenol-chloroform. The DNA was analyzed by qPCR using primers that were specific for regions spanning the Stat3-binding sites in the promoters of BCL-2, VEGF and SOCS3.
Human STAT3-SH2 expression and purification
Recombinant human STAT3-SH2 fusion protein was expressed in E. coli BL21 (DE3). Typically, BL21 (DE3) was transformed with pGEX4T-1-STAT3-SH2 plasmid. When the OD value reached about 0.6, the E. coli was transferred to 22 °C and 0.5 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) was added. The soluble protein was obtained by sonication and centrifugation. The protein was incubated with Glutathione-Sepharose beads (GE), and then eluted with 20 mM reducing glutathione solution. The protein was further concentrated with ultrafiltration centrifuge tube (Millipore) and Protein concentration was determined. The protein was stained with coomassie brilliant blue.
Surface Plasmon Resonance (SPR) analysis
This analysis was conducted with a Biacore T200 instrument (GE Healthcare) with CM5 sensor chip (GE Healthcare). In order to capture STAT3-SH2 with GST tag, GST antibody was immobilized in parallel-flow channels of CM5 sensor chip by using GST Capture Kit (GE Healthcare) and amine coupling kit (GE Healthcare). STAT3-SH2 with GST tag was captured. To test TSN-binding of STAT3-SH2 protein, series concentrations of TSN were injected into the flow system. Experiments were conducted with PBS buffer, and the analyte was injected at the flow rate of 30 μl/min. The association time was 60 s and the dissociation time was 30 s. Since TSN was dissolved in PBS with 5% DMSO, solvent correction assay was performed to adjust the results.
Co-immunoprecipitation assay
After treatment, 143B cells were collected with NP40 cell lysis buffer. Equal amount of proteins was incubated with antibodies overnight at 4 °C. Immuoprecipitates were incubated with protein A/G agarose beads and non-specific proteins were washed away by wash buffer.
MTS cell viability assay
Cells (5 × 103 per well) were seeded into 96-well plates. Forty-eight hours post TSN exposure, Aqueous One solution were added. The OD values at 490 nm were acquired.
Tumor cell clonogenic assay
Cell clonogenic assay was conducted as previously described.3 Tumor cells were seeded into 6-well plate. After tumor cell attachment, different concentrations of TSN were added. On day 7, colonies were fixed, and stained with 0.1% crystal violet. The colony number was counted manually.
Apoptosis assay
Apoptosis was measured using the Apoptosis Detection Kit (BD Biosciences) as previously described.40
Conditional medium collection and Tube formation assay
When cell density (143B cells) reached to 90%, the growth medium was replaced by serum-free medium containing indicted concentrations of TSN, and cells were incubated for 24 h at 37 °C. Conditional medium was subsequently collected and stored at −80 °C for subsequent tube formation assay. HUVECs (1 × 104) were seeded on Matrigel with conditional medium. After incubation for 8 h, photomicrographs were acquired and the lengths of tubular structures were quantified by Image-Pro Plus 6.0 software (Media Cybernetics, Bethesda, MA, USA).
Invasion assay
Invasion assay was conducted as described previously.41 In brief, after 12 h pretreatment with TSN, a total of 5 × 104 cells (for 143B) or 10 × 104 cells (for MG63.2) were seeded in the upper chamber. Different concentrations of TSN were added to both chambers. Invaded cells were stained with 0.1% crystal violet. And the number of invaded cells was counted manually.
Immunohistochemistry (IHC) analysis
IHC analysis was performed as previously reported.40 Sections (4 μm) were stained with indicted antibodies. Images were obtained with Leica microscope (Leica, DM4000b).
Three-dimensional (3D) on-top assay
3D on-top assay was performed as previously described.38 Briefly, Matrigel was added into a 48-well plate. 143B cells were resuspended in DMEM with 10% Matrigel as well as indicated TSN and then seeded on solidified Matrigel. The on-top mixture was changed every other day.
Fluorescent-gelatin degradation assay
This assay was performed as previously reported.42 In brief, cells was pretreated with TSN for 24 h, and then seeded on each coverslip (precoated with 1:9 0.1% fluorescein isothiocyante (FITC)-gelatin (Invitrogen): 0.2% porcine gelatin) with different concentrations of TSN. After 12 h, immunofluorescence analysis was performed. Gelatin degradation was quantified using Image-Pro Plus 6.0 software.
Animal model
All animal care and experimental studies were performed according to the guidelines and approval of the Animal Investigation Committee of the Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine. Male BALB/c athymic nude mice (6~8 weeks) and BALB/c mice (6~8 weeks) were bred and maintained at the animal experimental center in Shanghai General Hospital.
For osteosarcoma (143B) xenograft model, 143B tumor cells (1 × 106) were suspended in sterile PBS (20 μl) and injecteded into the medullary cavity of tibia of each mouse. After 7 days, three groups (n=6 per group) were randomly divided and received i.p. injection of TSN (1 mg/kg per 2 days and 2 mg/kg per 2 days) as compared with mice injected with DMSO (control group). After 24 days, all mice were executed. The posterior limb with tumors and lungs were finely excised for further study. Tumor weight was measured and lung metastasis nodules numbers were counted by three individuals who do not have personal biases with the current experiment. Another independent animal experiment (n=8 per group) was performed to determine whether TSN prolongs the survival of mice.
The patient-derived human osteosarcoma xenografts (PDXs) animal model was conducted according to previously described procedures.43 Briefly, seven pairs of osteosarcoma specimens and the corresponding non-tumor adjacent tissues were obtained from patients who had undergone a surgical osteosarcoma excision at Shanghai General Hospital of Shanghai Jiao Tong University, school of medicine. Fresh tumor tissue was sterilely incised into ~3 mm3 pieces, followed by s.c. into the flank of nude mice. The remaining tumor tissues were stored in liquid nitrogen. When the tumors have successfully engrafted, tumor samples were passaged into the third generation of nude mice for the following studies. On day 7, the mice were allocated into three groups (n=8) and recieved i.p. injection of TSN (1 mg/kg/2 days and 2 mg/kg/2 days) as compared with mice injected with DMSO (control group). Tumor volume was measured by a digital caliper in every week. Tumor volume was determined using the following formula: (length × width2) × 0.52. After 35 days, all the mice were killed. The tumors were removed and prepared for western blot and immunohistochemistry analyses.
H&E staining
Brain, heart, lung, kidney, livers and spleen were fixed overnight and paraffin-embedded. The sections were subjected to H&E staining.
Statistical analysis
The sample size was used to make sure adequate statistical power (≥ 80%), according to formal power calculation. The data are presented as mean±s.d. A Student’s t-test was used to compare two groups (P<0.05 was considered statistically significant) unless otherwise indicated. All experiments were performed at least three times except animal models. The variance per assay was similar between the groups statistically compared. No data points in our study were excluded.
References
Chun J, Li RJ, Cheng MS, Kim YS . Alantolactone selectively suppresses STAT3 activation and exhibits potent anticancer activity in MDA-MB-231 cells. Cancer Lett 2015; 357: 393–403.
Devarajan E, Huang S . STAT3 as a central regulator of tumor metastases. Curr Mol Med 2009; 9: 626–633.
Zhang T, Li S, Li J, Yin F, Hua Y, Wang Z et al. Natural product pectolinarigenin inhibits osteosarcoma growth and metastasis via SHP-1-mediated STAT3 signaling inhibition. Cell Death Dis 2016; 7: e2421.
Yu H, Jove R . The STATs of cancer—new molecular targets come of age. Nat Rev Cancer 2004; 4: 97–105.
Real PJ, Sierra A, De Juan A, Segovia JC, Lopez-Vega JM, Fernandez-Luna JL . Resistance to chemotherapy via Stat3-dependent overexpression of Bcl-2 in metastatic breast cancer cells. Oncogene 2002; 21: 7611–7618.
Zhang X, Sun Y, Pireddu R, Yang H, Urlam MK, Lawrence HR et al. A novel inhibitor of STAT3 homodimerization selectively suppresses STAT3 activity and malignant transformation. Cancer Res 2013; 73: 1922–1933.
Darnell JE Jr, Kerr IM, Stark GR . Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 1994; 264: 1415–1421.
Yu H, Pardoll D, Jove R . STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer 2009; 9: 798–809.
Bharadwaj U, Eckols TK, Kolosov M, Kasembeli MM, Adam A, Torres D et al. Drug-repositioning screening identified piperlongumine as a direct STAT3 inhibitor with potent activity against breast cancer. Oncogene 2015; 34: 1341–1353.
Zhang X, Yue P, Fletcher S, Zhao W, Gunning PT, Turkson J . A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochem Pharmacol 2010; 79: 1398–1409.
Hayakawa F, Sugimoto K, Harada Y, Hashimoto N, Ohi N, Kurahashi S et al. A novel STAT inhibitor, OPB-31121, has a significant antitumor effect on leukemia with STAT-addictive oncokinases. Blood Cancer J 2013; 3: e166.
Picci P . Osteosarcoma (osteogenic sarcoma). Orphanet J Rare Dis 2007; 2: 6.
Isakoff MS, Bielack SS, Meltzer P, Gorlick R . Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol 2015; 33: 3029–3035.
Fossey SL, Liao AT, McCleese JK, Bear MD, Lin J, Li PK et al. Characterization of STAT3 activation and expression in canine and human osteosarcoma. BMC Cancer 2009; 9: 81.
Ryu K, Choy E, Yang C, Susa M, Hornicek FJ, Mankin H et al. Activation of signal transducer and activator of transcription 3 (Stat3) pathway in osteosarcoma cells and overexpression of phosphorylated-Stat3 correlates with poor prognosis. J Orthop Res 2010; 28: 971–978.
Butler MS . Natural products to drugs: natural product-derived compounds in clinical trials. Nat Prod Rep 2008; 25: 475–516.
Bharti AC, Donato N, Aggarwal BB . Curcumin (diferuloylmethane) inhibits constitutive and IL-6-inducible STAT3 phosphorylation in human multiple myeloma cells. J Immunol 2003; 171: 3863–3871.
Kotha A, Sekharam M, Cilenti L, Siddiquee K, Khaled A, Zervos AS et al. Resveratrol inhibits Src and Stat3 signaling and induces the apoptosis of malignant cells containing activated Stat3 protein. Mol Cancer Ther 2006; 5: 621–629.
Cao L, Qu D, Wang H, Zhang S, Jia C, Shi Z et al. Toosendanin exerts an anti-cancer effect in glioblastoma by inducing estrogen receptor beta- and p53-mediated apoptosis. Int J Mol Sci 2016; 17: 1928.
Nair RR, Tolentino JH, Hazlehurst LA . Role of STAT3 in transformation and drug resistance in CML. Front Oncol 2012; 2: 30.
Hsieh FC, Cheng G, Lin J . Evaluation of potential Stat3-regulated genes in human breast cancer. Biochem Biophys Res Commun 2005; 335: 292–299.
Sakai I, Takeuchi K, Yamauchi H, Narumi H, Fujita S . Constitutive expression of SOCS3 confers resistance to IFN-alpha in chronic myelogenous leukemia cells. Blood 2002; 100: 2926–2931.
Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JE Jr, Yancopoulos GD . Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science 1995; 267: 1349–1353.
Folkman J . Role of angiogenesis in tumor growth and metastasis. Semin Oncol 2002; 29: 15–18.
Nicolson GL . Organ specificity of tumor metastasis: role of preferential adhesion, invasion and growth of malignant cells at specific secondary sites. Cancer Metastasis Rev 1988; 7: 143–188.
Nakajima M, Chop AM . Tumor invasion and extracellular matrix degradative enzymes: regulation of activity by organ factors. Semin Cancer Biol 1991; 2: 115–127.
Kang Y, Massague J . Epithelial-mesenchymal transitions: twist in development and metastasis. Cell 2004; 118: 277–279.
Malaney P, Nicosia SV, Dave V . One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer Lett 2014; 344: 1–12.
Marangoni E, Vincent-Salomon A, Auger N, Degeorges A, Assayag F, de Cremoux P et al. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin Cancer Res 2007; 13: 3989–3998.
Haan S, Hemmann U, Hassiepen U, Schaper F, Schneider-Mergener J, Wollmer A et al. Characterization and binding specificity of the monomeric STAT3-SH2 domain. J Biol Chem 1999; 274: 1342–1348.
Hao W, Hu Y, Niu C, Huang X, Chang CP, Gibbons J et al. Discovery of the catechol structural moiety as a Stat3 SH2 domain inhibitor by virtual screening. Bioorg Med Chem Lett 2008; 18: 4988–4992.
Ferrara N . VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer 2002; 2: 795–803.
Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene 2002; 21: 2000–2008.
Valastyan S, Weinberg RA . Tumor metastasis: molecular insights and evolving paradigms. Cell 2011; 147: 275–292.
Xiong H, Hong J, Du W, Lin YW, Ren LL, Wang YC et al. Roles of STAT3 and ZEB1 proteins in E-cadherin down-regulation and human colorectal cancer epithelial-mesenchymal transition. J Biol Chem 2012; 287: 5819–5832.
Lu X, Ji C, Tong W, Lian X, Wu Y, Fan X et al. Integrated analysis of microRNA and mRNA expression profiles highlights the complex and dynamic behavior of toosendanin-induced liver injury in mice. Sci Rep 2016; 6: 34225.
Su Y, Luo X, He BC, Wang Y, Chen L, Zuo GW et al. Establishment and characterization of a new highly metastatic human osteosarcoma cell line. Clin Exp Metastasis 2009; 26: 599–610.
Li J, Zhang T, Yang F, He Y, Dai F, Gao D et al. Inhibition of breast cancer progression by a novel histone deacetylase inhibitor, LW479, by down-regulating EGFR expression. Br J Pharmacol 2015; 172: 3817–3830.
Wang J, Zhang L, Chen G, Zhang J, Li Z, Lu W et al. Small molecule 1'-acetoxychavicol acetate suppresses breast tumor metastasis by regulating the SHP-1/STAT3/MMPs signaling pathway. Breast Cancer Res Treat 2014; 148: 279–289.
Zhang T, Chen Y, Li J, Yang F, Wu H, Dai F et al. Antitumor action of a novel histone deacetylase inhibitor, YF479, in breast cancer. Neoplasia 2014; 16: 665–677.
Zhang T, Li J, Dong Y, Zhai D, Lai L, Dai F et al. Cucurbitacin E inhibits breast tumor metastasis by suppressing cell migration and invasion. Breast Cancer Res Treat 2012; 135: 445–458.
Eckert MA, Lwin TM, Chang AT, Kim J, Danis E, Ohno-Machado L et al. Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell 2011; 19: 372–386.
Zhang H, Cohen AL, Krishnakumar S, Wapnir IL, Veeriah S, Deng G et al. Patient-derived xenografts of triple-negative breast cancer reproduce molecular features of patient tumors and respond to mTOR inhibition. Breast Cancer Res 2014; 16: R36.
Acknowledgements
This work was supported by NSFC (81502604); Shanghai Science and Technology Commission (14140904000); Doctoral Innovation Fund of Shanghai Jiaotong University School of Medicine (No. BXJ201732) and Research Grant from Shanghai Hospital Development Center (SHDC12013107).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Zhang, T., Li, J., Yin, F. et al. Toosendanin demonstrates promising antitumor efficacy in osteosarcoma by targeting STAT3. Oncogene 36, 6627–6639 (2017). https://doi.org/10.1038/onc.2017.270
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.270
This article is cited by
-
Toosendanin targeting eEF2 impedes Topoisomerase I & II protein translation to suppress esophageal squamous cell carcinoma growth
Journal of Experimental & Clinical Cancer Research (2023)
-
Traditional Chinese medicine for colorectal cancer treatment: potential targets and mechanisms of action
Chinese Medicine (2023)
-
Antitumor effect of toosendanin on oral squamous cell carcinoma via suppression of p-STAT3
BMC Oral Health (2023)
-
Retraction Note: Genipin suppression of growth and metastasis in hepatocellular carcinoma through blocking activation of STAT-3
Journal of Experimental & Clinical Cancer Research (2022)
-
Toosendanin, a late-stage autophagy inhibitor, sensitizes triple-negative breast cancer to irinotecan chemotherapy
Chinese Medicine (2022)