Abstract
Anti-EGFR antibodies are effective in therapies for late-stage colorectal cancer (CRC); however, many tumours are unresponsive or develop resistance. We performed genomic analysis of intrinsic and acquired resistance to anti-EGFR therapy in prospectively collected tumour samples from 25 CRC patients receiving cetuximab (an EGFR inhibitor). Of 25 CRC patients, 13 displayed intrinsic resistance to cetuximab; 12 were intrinsically sensitive. We obtained six re-biopsy samples at acquired resistance from the intrinsically sensitive patients. NCOA4–RET and LMNA–NTRK1 fusions and NRG1 and GNAS amplifications were found in intrinsic-resistant patients. In cetuximab-sensitive patients, we found KRAS K117N and A146T mutations in addition to BRAF V600E, AKT1 E17K, PIK3CA E542K, and FGFR1 or ERBB2 amplifications. The comparison between baseline and acquired-resistant tumours revealed an extreme shift in variant allele frequency of somatic variants, suggesting that cetuximab exposure dramatically selected for rare resistant subclones that were initially undetectable. There was also an increase in epithelial-to-mesenchymal transition at acquired resistance, with a reduction in the immune infiltrate. Furthermore, characterization of an acquired-resistant, patient-derived cell line showed that PI3K/mTOR inhibition could rescue cetuximab resistance. Thus, we uncovered novel genomic alterations that elucidate the mechanisms of sensitivity and resistance to anti-EGFR therapy in metastatic CRC patients.
Similar content being viewed by others
Introduction
Cetuximab-based chemotherapy has demonstrated survival benefit in patients with metastatic colorectal cancers (CRCs) over the past decade1,2,3,4. Cetuximab binds to the extracellular domain of the epidermal growth factor receptor (EGFR), which inhibits the RAS–RAF–mitogen-activated protein kinase 1 (MAPK1) and the v-akt murine thymoma viral oncogene homolog 1 (AKT1) axis, the pathways involved in cell proliferation, cell survival and tumour invasion5.
In approximately 40% of CRC patients, tumours harbour mutations in the v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS), mainly in codons 12, 13, and 616,7. KRAS mutations are the key negative predictive factors for cetuximab-based treatment in mCRC patients8,9. Although patients with KRAS wild-type (wt) CRC tumours are known to be responsive to cetuximab-based treatment, up to 65% of patients with KRAS wt tumours are resistant to anti-EGFR monoclonal antibodies10. Aberrations in other effectors of the EGFR signalling cascade (PIK3CA, PTEN, and NRAS) have been suggested to affect the primary response to cetuximab sensitivity11,12,13. In addition, MET amplification has been detected in CRC patients who initially responded to cetuximab but eventually acquired resistance14; however, MET amplification occurs only in 1% of CRC patients. Limited progress has been made in understanding the mechanism of resistance to cetuximab, particularly in patients who initially respond to cetuximab but acquire resistance during cetuximab-based chemotherapy.
In this study, we aimed to evaluate intrinsic and acquired resistance to anti-EGFR therapy in prospectively collected tumour samples of KRAS wt metastatic CRC (mCRC) patients who were administered cetuximab-containing regimens in real-world clinical care. We also attempted to obtain tumour tissues at tumour progression to investigate the genomic aberrations responsible for acquired resistance. Finally, we established patient-derived tumour cells from the tissues at the time of acquired resistance to cetuximab to explore alternative treatment regimens for these patients.
Results
Patient cohort
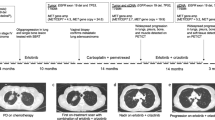
Genomic profiling was performed on 25 metastatic CRC patients treated with cetuximab-based chemotherapy (Table 1). All baseline tumours used for sequencing were from the primary tumour site. DNA and RNA were extracted for whole-exome and transcriptome sequencing, as well as copy number (CN) analysis using genotype arrays (Table S1). Fourteen patients were administered first-line cetuximab/FOLFIRI or cetuximab/FOLFOX for metastatic disease, and 11 patients were administered cetuximab/irinotecan as a salvage treatment. Patients who showed stable disease (SD) or progressive disease (PD) following cetuximab treatment were categorized as ‘intrinsic-resistant’, and patients with complete response (CR) or partial response (PR) were categorized as ‘intrinsic-sensitive’ (Fig. 1a). Moreover, among patients showing intrinsic sensitivity to cetuximab, those who developed resistance to cetuximab during cetuximab-based treatment or within 2 months following the completion of cetuximab-based treatment were defined as ‘acquired-resistant’. In the first-line setting (n = 14), eight (57.1%) patients achieved a confirmed PR (Fig. 1b). Of the 11 patients administered irinotecan/cetuximab as a salvage treatment, four (36.4%) achieved a PR (Fig. 1b). Of these 12 cetuximab-sensitive patients (eight in the first-line setting and four in the salvage setting), we successfully obtained re-biopsies at the time of acquired resistance in six patients (blue stars; four in the first-line setting and two in the salvage setting; Fig. 1b). The re-biopsy sites at acquired resistance following the initial response to cetuximab were as follows: colon, n = 2; peritoneal seeding, n = 2; bone, n = 1; and liver, n = 1.
Clinical response to cetuximab. (a) Representative computed tomography scans from cetuximab intrinsic-sensitive and acquired-resistant colorectal cancer (CRC) patients. (b) Horizontal bar plots represent time (months) for which patients were on cetuximab treatment until progressive disease (PD) (black dots) for first-line cetuximab-based chemotherapy (n = 14) (top) or salvage cetuximab/irinotecan chemotherapy (n = 11) (bottom). Orange, blue, and grey bars indicate partial response (PR), stable disease (SD), and PD, respectively. Blue stars indicate successful re-biopsy in patients who achieved PR and then developed acquired resistance. Vertical waterfall bar plots in the right panels show the percent change in tumour size (y-axis) from baseline during cetuximab treatment. As defined by RECIST criteria, patients who achieved a shrinkage in tumour size of >30% were classified as PR and ‘intrinsic sensitive’ for our study.
Intrinsic-resistant tumours
The genomic landscape of the intrinsic-sensitive, intrinsic-resistant, and acquired-resistant tumours using exome, transcriptome, and CN analysis is shown in Fig. 2. Genetic alterations (Fig. 2a,b) in RAS/RAF pathway regulators were frequently observed in intrinsic-resistant tumours. Although all patients were selected based on direct sequencing of KRAS at the time of cetuximab administration, whole-exome sequencing identified four additional patients with KRAS mutations (two with G12V and once each with G13D and Q61H). An NRAS G12D mutant tumour also showed cetuximab resistance. Nearly all tumours profiled showed a low somatic mutation burden (0.0–3.9 non-silent Mut/Mb), which is consistent with our previous classification of all patient tumours as microsatellite stable. Notably, the two exceptions with high mutation burden (patient #3080, 29.5 Mut/Mb; patient #73, 17.8 Mut/Mb) showed cetuximab resistance (Fig. 2a, top). Consistent with the results of recent studies, we also detected point mutations in ERBB2 and PDGFRA that likely confer cetuximab resistance15,16. The PDGFRA A978T variant is near the PDGFRA kinase domain and is close to the R981H mutation, which has been shown to confer cetuximab resistance in an earlier study15. In that same study, a xenograft model carrying the PDGFRA R981H mutation treated with imatinib in combination with cetuximab showed increased efficacy, suggesting that PDGFRA kinase inhibitors might be useful for treating cetuximab-resistant CRC. One somatic EGFR mutation, D522Y, was also identified in the EGFR extracellular domain of an intrinsic-resistant tumour; however, residue 522 is not in the EGFR cetuximab-binding domain. Its impact on cetuximab affinity17 requires further validation.
Landscape of somatic variants and gene set expression signatures in sensitive, resistant, and acquired-resistant tumours. (a,b) Somatic variants identified in genes grouped by molecular pathways. MUT, non-synonymous single nucleotide or small indel mutation; FUS, gene fusion; AMP, copy number amplification (CN ≥4). The top bar chart shows the tumour mutation burden of non-synonymous somatic variants. Amino acid change is shown for a subset of genes in (b). (c) Hierarchical clustering of gene set expression signatures for a panel of published molecular signatures (see Methods). The Z score represents both the magnitude and relative direction of a signature’s expression. Baseline tumour (T), acquired-resistant tumour (RT).
Using RNA sequencing data, we also identified two in-frame gene fusions (Tables S2, S3, and Fig. 3), including NCOA4–RET and LMNA–NTRK1 fusions. These fusions were both confirmed to be tumour-specific (Fig. 3b,f) and preserved the tyrosine kinase domain in the 3′ fusion partners RET and NTRK1, respectively (Fig. 3a,d,e,h). Expression analysis further revealed the dramatic upregulation of RET and NTRK1 fusion transcripts in the tumours relative to their matched normal tissue (Fig. 3c,g). While NTRK1 fusions have been reported in cetuximab-resistant patients18,19, this is the first clinical report demonstrating RET fusion as the putative cause of cetuximab resistance. RTK fusions, including RET and NTRK1 fusions, have been identified in ~0.4% of CRC patients16.
Characterization of NCOA4–RET and LMNA–NTRK1 gene fusions. (a,e) Schematic depiction of the gene fusion detected by RNA-Seq, indicating the genomic position, resulting fusion mRNA, and predicted protein consequences (with key protein domains indicated). (b,f) RT-PCR of RNA from the tumour or adjacent normal tissue with primers upstream and downstream of the gene fusion breakpoint. NTC, no template control. Red arrows indicate the size of the expected PCR product. (c,g) Overexpression of the 3′ fusion gene partner in the tumour relative to the adjacent normal tissue. RPM, reads per million. (d,h) Sanger sequencing of the RT-PCR product from (b) and (f). The red arrowheads indicate the position of the fusion junction and in-frame amino acid sequence.
Genome-wide CN analysis of the 13 intrinsic-resistant tumours revealed NRG1 and GNAS amplifications in CRC tumours without any aberrations in RAS/RAF pathway genes (Fig. 2a). Neuregulin 1 (NRG1; also known as heregulin), encodes an EGF-like signalling molecule that binds and activates ERBB2/3 heterodimers. The high CN of NRG1 as a resistance marker is consistent with previous findings of higher NRG1 protein and RNA expression levels corresponding to cetuximab resistance13,20. GNAS encodes the Gs-alpha subunit of G-proteins, and GNAS amplification or point mutations have been identified in approximately 10% of CRC tumours, often coinciding with KRAS mutations21. GNAS gain-of-function variants result in elevated cAMP and ERK/MAPK activation22. GNAS was also predicted to play a role in resistance to BRAF + EGFR inhibition in BRAF V600E mutant CRC23. GNAS amplification in patient #53 correlates with >6-fold increased RNA expression.
Furthermore, to explore whether changes in the expression of known molecular pathways contribute to cetuximab resistance, we performed gene set enrichment analysis (Figs 2c, S1). We identified no consistently up- or downregulated pathways for differentiating sensitive from resistant tumours in well-characterized pathways in CRC. Overall, our genomic analyses identified potential resistance-causing alterations in 11 of the 13 intrinsic-resistant tumours. These variants appear to bypass the anti-EGFR blockade by activating alterations in RAS/RAF pathway genes.
Intrinsic-sensitive tumours
For cetuximab-sensitive tumours, there were no major aberrations in the RAS pathway in any of the 12 tumours (Fig. 2a,b). Two patients showed KRAS K117N and A146T mutations, respectively, which are in KRAS exon 4 rather than in the more dominant exon 2 (G12/13)24,25. We also found other RTK/RAF and PI3K/AKT pathway mutations in the cetuximab-sensitive cohort, including BRAF V600E (n = 1), AKT1 E17K (n = 2), PIK3CA E542K (n = 1), FGFR1 amplification (n = 1), and ERBB2 amplification (n = 2). In patient #5804, in addition to an ERBB2 amplification in primary and metastatic lesions, we also discovered an ERBB2–MED24 fusion present only in liver metastasis (Table S2 and Fig. S2). The fusion lacks the ERBB2 kinase domain, and expression analysis indicates that it did not contribute to ERBB2 expression (data not shown), leading us to conclude that it might not be a driver mutation. Extended analysis of ERBB2 fusion transcripts in TCGA RNA-Seq data shows a close association of ERBB2 fusions with high ERBB2 CN, possibly a result of secondary passenger events in the amplification process (Table S4).
Acquired-resistant tumours
Among the 12 cetuximab-sensitive patients, 11 developed acquired resistance following the response to cetuximab, and we obtained follow-up resistant tumour specimens from six patients (Fig. 1b). We successfully profiled all six acquired-resistant tumours using exome sequencing, but due to low tissue volume and quality, we were only able to analyse five for CN variants and three with RNA-Seq (Table S1). We compared the variant and CN profiles between the pre-treated baseline and re-biopsied tumours as shown in Fig. 2. Previously described cetuximab resistance-related genomic aberrations were not found in the acquired-resistant tumours, and novel variants did not immediately suggest any apparent resistance mechanisms.
We plotted and compared the variant allele fraction (VAF) of silent and non-silent variants in six pre- and post-treatment paired samples (Fig. 4a). The comparison revealed both an extreme loss (variants found along the x-axis, VAF = 0 for acquired-resistant tumour) and simultaneous gain (variants found along the y-axis, VAF = 0 for baseline tumour) of somatic variants in acquired-resistant tumours at the time of disease progression. Of note, patient #GJG showed no gain of non-silent acquired-resistant variants. These results suggest that cetuximab exposure dramatically selected for rare resistant subclones of the baseline tumour that were undetectable at the start of the treatment. These observations were further validated by CN array B-allele frequency (BAF) analysis in the re-biopsied resistant samples (Fig. 4b). The BAF plots showed many loci with BAF imbalance in the baseline-sensitive tumours, which were ‘lost’ in the acquired-resistant tumours. For example, chromosome 10 in patient #GJG showed BAF skewing in the sensitive tumour, which was no longer present in the acquired-resistant tumour. Because tumour evolution is unlikely to restore BAF imbalances as the tumour progresses, we concluded that the acquired-resistant tumour was likely derived from a minor subclone that originally had normal BAF balance at the chromosome 10 locus prior to cetuximab treatment. Patient #4612 showed the most dramatic changes in BAF, which is also consistent with the shift observed in its non-silent VAF.
Variant and copy number changes in sensitive and acquired-resistant tumour pairs. (a) Variant allele frequency (VAF) of somatic mutations detected in baseline (T) versus acquired-resistant (RT) tumours. Variants were grouped as non-silent (blue) and silent (black) based on the predicted impact on protein sequence. (b) B-allele frequency (BAF) plots show regions of genomic imbalance from copy number variants for each chromosome (1–22) of T and RT.
To evaluate whether clonal selection of acquired-resistant tumours would be reflected in the activation of specific CRC-related transcription pathways, we looked for enrichment scores for gene expression pathways between baseline and acquired-resistant tumour pairs (pts. #JJ, #4612, #PYM) (Fig. 5a). Patient #4612 showed enrichment for several molecular pathways, including an increase in epithelial-to-mesenchymal transition (EMT), with a reduction in the immune infiltrate. To explore these findings further, we compared the expression of all genes constituting the EMT signature and found a clear upregulation of mesenchymal markers compared to epithelial markers in the acquired-resistant tumour from patient #4612 but not patient #JJ or patient #PYM (Fig. 5b). Using the ESTIMATE tool to quantify the immune and stromal infiltration, we also confirmed that patient #4612 had a dramatic reduction in immune and stromal scores (Fig. 5c)26. Additional signature analyses of EMT and immune and metabolic pathways in patient #4612 are depicted in Supplementary Fig. S3. Patient #JJ also demonstrated a drop in the stromal infiltration score but instead showed an increase in the immune infiltration score at resistance (Fig. 5c). In addition, patient #JJ also showed an increase in MYC signalling in his parental versus acquired-resistant samples (Fig. 5a). No clear change was observed in the expression signatures for patient #PYM. These results suggest that multiple mechanisms are involved in clonal evolution under selective pressure by cetuximab.
Gene expression signatures in sensitive and acquired-resistant tumour pairs. Analysis of RNA expression signatures and enrichment scores in sensitive versus acquired-resistant tumour pairs. Baseline tumour (T), acquired-resistant tumour (RT). (a) Pathway enrichment Z-scores plotted for each molecular pathway expression signature. The green line represents a smooth local regression LOESS curve, and grey shading highlights the 95% confidence interval around the curve. Pathways with the highest enrichment Z-scores in each panel are labelled. Red arrows highlight pathways of interest. (b) Expression (log2) of individual genes that comprise the epithelial (E.EMT) and mesenchymal (M.EMT) gene expression signature. A diagonal line is plotted for comparison. The EMT signature genes showing >4-fold change difference of expression between baseline tumour and acquired-resistant tumour are labelled. (c) Gene expression signatures for stromal and immune cell tumour infiltration were quantified using the ESTIMATE tool (see Methods). Orange bars represent scores for immune cell infiltration and grey bars, scores for stromal cell infiltration.
In vitro analysis of an acquired-resistant, patient-derived cell line
To investigate the development of a treatment strategy for overcoming the acquired resistance to EGFR-targeted therapy, we established a cetuximab-resistant, patient-derived cell (PDC) line from peritoneal seeding tissue of patient #4612 at acquired resistance. This 60-year old female patient initially had KRAS wt, NRAS wt, and TP53 mt colorectal mucinous adenocarcinoma with peritoneal seeding and showed a dramatic response to cetuximab for 5 months; however, during cetuximab treatment, she was found to have peritoneal seeding (Fig. 6a).
Sensitivity of a patient-derived cell line from patient #4612 to PI3K/mTOR inhibition. (a) Computed tomography scans for patient #4612 at baseline and following 3 and 12 cycles of cetuximab treatment. A patient-derived cell (PDC) line was cultured from the acquired-resistant tumour (malignant ascites, which developed following 12 cycles). (b) Dose-dependent inhibition of PDC cell proliferation by 5-day treatments with cetuximab (EGFR), erlotinib (EGFR), GDC-0994 (Erk1/2), BEZ-235 (dual PI3K/mTOR), AZD-8055 (mTOR), and BKM-120 (pan-PI3K). Results from four independent experiments are shown. The IC50 values of AZD-8055, BEZ-235, and BKM-120 are 0.02, 0.047, and 0.971 µM, respectively. (c) Western blot expression of basal and 24 h treatments of PDC. Cetuximab concentration, 75 µg/mL; other compounds, 1 µM. The experiments were repeated thrice.
To confirm that the PDC line originated from the acquired-resistant tumour, we measured the expression of EMT markers and performed Sanger sequencing of two acquired-resistant mutations. We correspondingly observed a loss of E-cadherin expression and gain of N-cadherin, vimentin, and AXL expression in the acquired-resistant tumour for patient #4612 (Table S5). Similarly, we confirmed the presence of the acquired variants GTF2H5 and SLC17A1 (Table S6).
Cell proliferation assays using the PDC showed resistance to EGFR inhibition with cetuximab or erlotinib as expected (Fig. 6b). We next tested whether the cetuximab-resistant PDC would be sensitive to the inhibition of the MEK/ERK pathway, which has been suggested as a cetuximab resistance mechanism in previous cell line and animal studies27,28. However, the Erk1/2 inhibitor GDC-0994 did not suppress the growth of the PDC line (Fig. 6b). Further analysis of the acquired mutations and RNA expression in the acquired-resistant tumour for patient #4612 revealed both an acquired IRS1 P313S variant (Fig. 2b), as well as a ~5-fold decrease in the expression of the PI3K regulatory subunit PIK3R1, leading us to hypothesize that the PI3K/mTOR pathways might be dysregulated in the resistant tumour. Although canonical PI3K/mTOR expression signatures were not considerably altered in our gene signature analysis closer investigation of individual gene expression revealed greater expression shifts relative to baseline for patient #4612 (Fig. S4), which could impact protein-level pathway activation that is not captured by the canonical RNA expression signatures. Indeed, the PDCs were sensitive to dual PI3K/mTOR (BEZ-235), mTOR (AZD-8055), and to a lesser degree, pan-PI3K (BKM-120) treatments. Immunoblot assays for downstream pathway activation also revealed that dual PI3K/mTOR (BEZ-235) and mTOR (AZD-8055) treatments potently inhibited the phosphorylation of S6RPS240/244 and 4E-BP1S65/101 (Fig. 6c), which was concordant with the inhibition of cell proliferation.
Discussion
The aim of this study was to evaluate the mechanisms underlying sensitivity and resistance to cetuximab-based chemotherapy in CRC patients. Given the complexity of EGFR crosstalk that affects sensitivity and resistance to anti-EGFR therapy in mCRC as previously described29, a simple genomic aberration or pathway may not explain the sensitivity or resistance to cetuximab in mCRC. Comparative genomic analyses suggested that most of the acquired-resistant tumours in our study resulted from a selective outgrowth of a minor clonal variant in the primary tumour that was not detectable at baseline by the standard NGS approach, as suggested earlier30. Detailed analysis of intrinsic-resistant tumours revealed several single-nucleotide variants in KRAS, NRAS, ERBB2, and PDGFRA, which likely confer resistance. Moreover, we identified NCOA4–RET and LMNA–NTRK1 fusions in two patients who were both refractory to cetuximab-based chemotherapy. The LMNA–NTRK1 fusion was recently identified as a potential target in cetuximab-resistant tumours18. The largest global survey of 17 RET fusion-positive mCRC patients demonstrated that all patients with rearranged RET had KRAS wt, and 10 of them had an NCOA4–RET fusion associated with significantly poorer survival compared to the RET fusion-negative KRAS wt patients31,32. This is the first study to detect an NCOA4–RET fusion in cetuximab-resistant KRAS wt CRC. The possibility that RET fusions drive resistance to EGFR inhibition is corroborated by recent studies showing that RET fusions confer resistance to osimertinib in non-small cell lung cancer patients33. Furthermore, the lung cancer cell line Lc2/ad, which carries a RET fusion, shows more resistance to erlotinib compared to SW48 cells, and direct targeting of RET inhibition in Lc2/ad cells can be bypassed by EGFR activation34,35.
Novel amplifications of NRG1 and GNAS may predict resistance to cetuximab (both found in KRAS wt, NRAS wt patients). In the RAS pathway, NRG1 and other EGFR ligands were previously found to play a major role in conferring primary cetuximab resistance in CRC pre-clinical models, although a correlation between NRG1 expression and NRG1 amplification was not reported36. The patient harbouring the NRG1 amplification (patient #3772) was a 76-year-old woman with poorly differentiated adenocarcinoma, who demonstrated progression after three cycles of cetuximab/irinotecan. NRGs are encoded by four individual genes (NRG1–4), and NRG1 generates six types of proteins (I–VI) in at least 31 isoforms (29). All isoforms contain an extracellular EGF-like domain that induces the activation of ERBB RTKs. Recently, an NRG1 fusion inducing ERBB3 activation was characterized in lung cancer37 and breast cancer38; however, NRG1 amplification has not been characterized in CRC.
In cetuximab-sensitive tumours, we identified several mutations in PI3K/AKT and RTK/RAS pathway genes, which would be expected to confer cetuximab resistance, including variants in BRAF, KRAS, PIK3CA, AKT1, FGFR1, and ERBB2. Our findings suggest that the presence of these specific variants alone is not sufficient to preclude an objective response to cetuximab. A recurrent AKT1 E17K hotspot mutation was found in two out of 25 patients in our cohort (8%). In one study, AKT1 was found to be mutated in only ~1.5% of TCGA CRCs39, suggesting that the mutation frequency is higher in Asian populations. Several of these putative resistance variants co-occurred in three cetuximab-sensitive tumours, indicating that some interplay between the variants may sensitize tumours to cetuximab.
Notably, patient #4612 acquired resistance to cetuximab and demonstrated several interesting genomic features at progression. First, the EMT signature was low at baseline, but was converted to a high EMT subtype at progression to cetuximab, which corresponds with recent findings showing that five of eight paired biopsies exhibited subtype switching to EMT at PD40. EMT has been associated with an invasive and metastatic phenotype, as well as drug resistance to both chemotherapy and molecularly targeted agents, while mechanisms to reverse EMT have been shown to re-sensitize some tumours to drug rechallenge41,42. Second, the PDC line established at acquired resistance from this patient was sensitive to dual PI3K/mTOR and mTOR inhibition. The re-biopsied tumour at resistance was PIK3CA wt but had low PIK3R1 expression and acquired an IRS1 P313S variant that was not present in the sensitive tumour. The PIK3R1 gene encodes the p85 regulatory subunit of PI3K, and IRS1 directly binds with p85 following IRS1 tyrosine phosphorylation43,44. IRS1 tyrosine phosphorylation is downregulated by the phosphorylation of multiple IRS1 serine residues in an mTOR-dependent manner. Proline-directed serine phosphorylation of human IRS1 Ser312, immediately adjacent to the IRS1 P313S variant, is a key serine residue for IRS1 feedback regulation43,44,45. Thus, low expression of p85 and an inability of IRS1 to respond to negative feedback signals could contribute to higher PI3K/mTOR activity and cetuximab resistance. It is unclear if EMT and acquired-resistant IRS1/PIK3R1 alterations are both contributing to cetuximab resistance in pt. #4612. EMT and ALK mutations were recently found to co-exist and separately contribute to resistance in the same crizotinib-resistant tumor lesions. Inhibition of the PI3K/mTOR pathway has also been previously shown to reverse expression of EMT markers, however, reversion of EMT alone does not directly account for blocking cell proliferation as demonstrated in recent studies that have pharmacologically reversed EMT but needed additional drug rechallenge to inhibit tumor growth. Therefore, we propose that sensitivity to PI3K/mTOR inhibitors is most likely due to IRS1/PIK3R1 acquired mutations rather than EMT reversion. Nonetheless, regardless of the specific resistance mechanism, our results clearly show that the patient-derived tumor cells are ‘oncogene addicted’ to PI3K/mTOR activation and therefore inhibition of this pathway is sufficient to overcome cetuximab resistance in this patient.
In summary, we show that NCOA4–RET and LMNA–NTRK1 fusions, along with NRG1 and GNAS amplifications, are potentially novel cetuximab-resistance (intrinsic) alterations in patients with KRAS wt CRC. Clonal selection in acquired resistance appears to be common with changes in CRC-related pathways and its microenvironment. We demonstrated that a PDC model derived from an acquired-resistant tumour was successfully suppressed by PI3K/mTOR inhibitors such as AZD-8055 or BEZ-235. The clinical benefit from these molecules should be evaluated in clinical trials, especially in a salvage setting with patients who have acquired resistance to cetuximab.
Methods
Patients and samples
Twenty-five mCRC patients administered treatment containing cetuximab were enrolled for the prospective collection of tumour samples. All study participants provided written informed consent. The study was approved by the institutional review board at Samsung Medical Center (IRB# 2013-10-014). The study was conducted in accordance with the Declaration of Helsinki and the Guidelines for Good Clinical Practice. Baseline tumour samples were collected before initiating cetuximab-based chemotherapy in all enrolled patients, and additional tumour samples were obtained at progression. All collected tumour samples were confirmed by our pathologists for tumour content and characteristics. All hematoxylin and eosin-stained slides were also reviewed. Patient age at diagnosis, sex, Eastern Cooperative Oncology Group performance status, number of involved organs, metastatic site, and chemotherapy data were collected.
Study procedure and statistical analysis
Biopsies were performed before cetuximab-based chemotherapy regimens. DNA and RNA were extracted for whole-exome and transcriptome sequencing, as well as CN analysis using genotype arrays. Response criteria were assessed every 2 months by computed tomography scanning and defined according to Response Evaluation Criteria in Solid Tumors (RECIST) 1.1.
Exome sequencing
Whole-exome sequencing was performed on an Illumina HiSeq. 2000 system (San Diego, CA, USA) using the SureSelect Human All Exon v4 (51 Mb) capture protocol (Agilent Technologies, Santa Clara, CA, USA). Paired-end sequencing with a read length of 100 base pairs and 100X average on-target coverage was conducted. Sequencing reads were mapped to the human genome (hg19) using the Burrows-Wheeler Aligner MEM algorithm. Variants were called using SAMtools24, Genome Analysis Toolkit (GATK-lite version)46, and FreeBayes47. Variants were filtered with a genotype Q score of ≥30, ≥3 variant reads, variant allele fraction of ≥0.05, and read coverage of ≥10 reads. Somatic mutations were called at positions with matched normal coverage of ≥10 reads, normal variant allele fraction of ≤0.05, normal variant allele fraction <2X of the tumour variant fraction, and no more than two pooled normal samples (pooling of all matched normal samples) with a variant allele fraction of ≥0.20.
CN analysis
DNA was prepared and loaded according to the manufacturer’s protocol on Affymetrix’s Genome-Wide Human SNP Array 6.0 (SNP6; Santa Clara, CA, USA). CEL files were used for CN analysis using three methods: (i) PICNIC47, for ploidy estimation; (ii) ASCAT48, for genome-wide visualization; and (iii) the Copy Number Inference Pipeline (CNIP) in GenePattern, for gene CNs. Three samples (5225AR, 4612S, and 7522S) failed to be completed using the PICNIC method. Candidate cancer genes included in the Sanger Cancer Census Gene list (http://cancer.sanger.ac.uk/census) with CN calls of ≥4 or <0.5 copies based on CNIP analysis were further confirmed using exome CN calls from CNVkit49. CN calls without matching support in SNP6 and exome data were not considered reliable.
RNA sequencing analysis
RNA sequencing of all samples was conducted in four data delivery batches. RNA-Seq was performed on an Illumina HiSeq. 2000 with the Illumina TruSeq RNA Sample Preparation Kit v2. Paired-end sequencing with a read length of 100 bp and targeted read depth of 50 million reads/sample was performed. Sequencing reads were mapped to the human reference genome (B37.p5 including all alternative contigs) using GSNAP (Wu et al., version 2013-11-27) with NCBI annotation as of December 2013. Read counts were generated against exons annotated in NCBI gene models and then summarized at the gene level to provide a single number/gene/sample using a custom perl script. Data were filtered to remove genes with fewer than 10 counts across 80% of the samples from the analysis. The resulting data were quantile-normalised and summarised across samples. Clustering analysis was carried out using all genes (21,344 genes) and genes with relatively high signals (10,346 genes, signals >10 in all samples). All samples were grouped into three batches based on clustering analysis results.
Data analysis was carried out on log2-transformed signals using a proc mixed procedure in SAS (SAS Institute, Cary, NC, USA). The statistical model included sensitivity as a fixed effect and batch as a random effect. Samples from the same subject were treated by repeated measurement using the covariance matrix option toep. A small number of genes used the ar(1) option if the model could not converge with toep. The results of comparisons of interest were then derived from the statistical modelling outputs. Fold changes and mean signals were converted back to count levels.
Fusion gene detection
BAM files generated by GSNAP (see above) were investigated for reads that had split alignments, identifying more than one gene either through normal splicing (indicated by an ‘N’ CIGAR) or through chimeric alignment (indicated by the ‘XT’ tag). Only fusions joining CDS in-frame and including at least 10 amino acids from both proteins were retained. Normal splicing reads for genes within 5 KB were discarded. Putative fusions previously found to occur in >1% of a large internal collection of non-cancer samples or in >10% of cancer samples were discarded as likely false-positives. Paired-end reads identifying putative gene fusions were also recorded. Potential fusions with ≥5 total reads (split and paired) were subjected to further analysis.
Fusion gene confirmation
RNA from tumour and normal tissues were used to produce cDNA using the SuperScript VILO MasterMix (Invitrogen, Carlsbad, CA, USA; #11755-050). A forward primer upstream of the fusion junction (previously identified from RNA-Seq data) and reverse primer downstream of the fusion junction were designed using the online program Primer3 for PCR and Sanger sequencing primers. The primer sequences are NCOA4_F1: GAGCCTGAGAAGCATAAAGATTCC, RET_R1: CCCATACAATTTGATGACATGTGG, LMNA_F2: CAAAGTGCGTGAGGAGTTTAAGG, NTRK1_R2, and CACTGAAGTATTGTGGGTTCTCG. PCR was carried out using Advantage®2 Polymerase Mix (Clontech, Mountain View, CA, USA; #639201) with Advantage 2 PCR buffer and cycled at 95 °C for 2 min; 35 cycles of 95 °C for 30 s; 65 °C for 30 s, 72 °C for 30 s; and a final extension at 72 °C for 10 min. PCR was also performed for the GAPDH gene as a control (GAPDH_F: ATCCCATCACCATCTTCCAG, GAPDH_R: CCATCACGCCACAGTTTCC). PCR products were purified with a Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA; #A9281). Sequencing PCR was performed using an ABI BigDye Terminator v3.1 cycle sequencing kit (Life Technologies, Carlsbad, CA, USA; #4337457). The resulting products were run on an ABI 3730xl DNA analyser. All sequences were visually analysed with Sequencher (Gene Codes Corp., Ann Arbor, MI, USA).
PCR and Sanger sequencing primers used to confirm the identity the PDC line were as follows: SLC17A1_F: TACCACTCAGCCAGTCAAATACC, SLC17A1_R: TCTGTGGTGACACTAGAAAGTTGC; GTF2H5_F: TTGTTAACACTTGAGGCAGAGAGG, GTF2H5_R: CCAAATTACAGCCAACTGTTAAAGC.
Gene set enrichment analysis
We performed gene set enrichment analysis using previously described signatures and well-characterized pathways in CRC50. These gene sets included canonical pathways, immune signatures, an immune and stromal cell admixture in tumour samples, and metabolic pathways. The signature score as an activation index was calculated using a combined Z score method51,52, which combines Z scores for each gene in a signature set for each sample. The Z score represents both the magnitude and relative direction of a signature’s expression. ESTIMATE R package was used to quantify immune and stromal cell infiltration26.
Patient-derived cell culture
Fresh tissue specimens were extensively washed with serum-free RPMI 1640, minced, and enzymatically dissociated for 2 h at 37 °C with agitation in serum-free RPMI 1640 containing 0.4 mg/mL collagenase (Gibco, Grand Island, NY, USA), 0.5 mg/mL dispase (Gibco), and 0.2 mg/mL DNase I (Roche, Basel, Switzerland), as described previously53. The cells were cultured in RPMI 1640 supplemented with 10% foetal bovine serum. Experiments were conducted within four passages after PDC derivation.
Cell proliferation assay
Cells were seeded at 5,000 cells per well in 100 μL of media in a 96-well plate, grown overnight, and then treated with different concentrations of drugs for 5 days prior to analysis using CellTiter-Glo (Promega; #G7572) according to the manufacturer’s instructions. All treatments were performed in duplicate, and four independent experiments were performed.
Immunoblot analysis
Cells were treated with 75 µg/mL cetuximab and 1 M erlotinib, GDC-0994, BEZ-235, AZD-8055, and BKM-120 for 24 h and then lysed with RIPA lysis buffer + Halt protease/phosphatase inhibitor cocktail. Protein concentration was measured using the BCA protein assay. Equal amounts of protein were separated on a 4–12% Bis-Tris Criterion polyacrylamide gel (Bio-Rad, Hercules, CA, USA) in MOPS buffer under reducing conditions and then transferred to a nitrocellulose membrane. The membranes were blocked with Odyssey blocking buffer in Tris-buffered saline and probed overnight at 4 °C with the indicated primary antibodies diluted in Odyssey blocking buffer + 0.2% Tween-20. The phosphor- and total EGFR, AKT, S6RP, 4E-BP1, and ERK antibodies were obtained from Cell Signaling Technology (Danvers, MA, USA), while the β-actin antibody was from Sigma (St. Louis, MO, USA). IRDye secondary antibodies were from Li-Cor, and a Li-Cor Odyssey scanner was used for visualization (Lincoln, NE, USA).
Real-time quantitative reverse transcription PCR
Real-time quantitative reverse transcription PCR (RT-qPCR) assays were performed using Taqman® probes for E-cadherin (CDH1; assay ID Hs01023895_m1), N-cadherin (CDH2; Hs00983056_m1), vimentin (Hs00958111_m1), and AXL (Hs01064444_m1). All reactions were performed in triplicate in 384-well plates following the manufacturer’s instructions and measured with a Quant Studio7 Flex (Thermo Fisher Scientific, Waltham, MA, USA). RT-qPCR analysis was performed using the comparative Ct (cycle threshold) method (2−ΔΔCt) using RNA18S5 as the control gene; the results are reported as the fold difference from matching normal tissue.
Data availability
The datasets generated during and/or analysed during the current study are available in the European Nucleotide Archive (ENA) repository, (Accession Number: PRJEB34338).
References
Douillard, J. Y. et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 28, 4697–4705 (2010).
Starling, N. & Cunningham, D. Cetuximab in previously treated colorectal cancer. Clin Colorectal Cancer 5(Suppl 1), S28–33 (2005).
Tabernero, J. et al. Phase II trial of cetuximab in combination with fluorouracil, leucovorin, and oxaliplatin in the first-line treatment of metastatic colorectal cancer. J Clin Oncol 25, 5225–5232 (2007).
Van Cutsem, E. et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 360, 1408–1417 (2009).
Bardelli, A. & Siena, S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol 28, 1254–1261 (2010).
Poehlmann, A. et al. K-ras mutation detection in colorectal cancer using the Pyrosequencing technique. Pathol Res Pract 203, 489–497 (2007).
Raponi, M., Winkler, H. & Dracopoli, N. C. KRAS mutations predict response to EGFR inhibitors. Curr Opin Pharmacol 8, 413–418 (2008).
De Roock, W. et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304, 1812–1820 (2010).
Karapetis, C. S. et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359, 1757–1765 (2008).
Allegra, C. J. et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 27, 2091–2096 (2009).
De Roock, W. et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. The Lancet. Oncology 11, 753–762 (2010).
Bertotti, A. et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer discovery 1, 508–523 (2011).
Yonesaka, K. et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Science translational medicine 3, 99ra86 (2011).
Bardelli, A. et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer discovery 3, 658–673 (2013).
Bertotti, A. et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature 526, 263–267 (2015).
Rankin, A. et al. Broad Detection of Alterations Predicted to Confer Lack of Benefit From EGFR Antibodies or Sensitivity to Targeted Therapy in Advanced Colorectal Cancer. The oncologist (2016).
Voigt, M. et al. Functional dissection of the epidermal growth factor receptor epitopes targeted by panitumumab and cetuximab. Neoplasia (New York, N.Y.) 14, 1023–1031 (2012).
Sartore-Bianchi, A. et al. Sensitivity to Entrectinib Associated With a Novel LMNA-NTRK1 Gene Fusion in Metastatic Colorectal Cancer. Journal of the National Cancer Institute 108 (2016).
Lee, S. J. et al. NTRK1 rearrangement in colorectal cancer patients: evidence for actionable target using patient-derived tumor cell line. Oncotarget 6, 39028–39035 (2015).
Zhang, L. et al. ERBB3/HER2 signaling promotes resistance to EGFR blockade in head and neck and colorectal cancer models. Molecular cancer therapeutics 13, 1345–1355 (2014).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery 2, 401–404 (2012).
Wilson, C. H., McIntyre, R. E., Arends, M. J. & Adams, D. J. The activating mutation R201C in GNAS promotes intestinal tumourigenesis in Apc(Min/+) mice through activation of Wnt and ERK1/2 MAPK pathways. Oncogene 29, 4567–4575 (2010).
Hong, D. S. et al. Phase IB Study of Vemurafenib in Combination with Irinotecan and Cetuximab in Patients with Metastatic Colorectal Cancer with BRAFV600E Mutation. Cancer discovery 6, 1352–1365 (2016).
Loupakis, F. et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. British journal of cancer 101, 715–721 (2009).
Sorich, M. J. et al. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: a meta-analysis of randomized, controlled trials. Annals of oncology: official journal of the European Society for Medical Oncology 26, 13–21 (2015).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 4, 2612 (2013).
Troiani, T. et al. Primary and acquired resistance of colorectal cancer cells to anti-EGFR antibodies converge on MEK/ERK pathway activation and can be overcome by combined MEK/EGFR inhibition. Clin Cancer Res 20, 3775–3786 (2014).
Misale, S. et al. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Science translational medicine 6, 224ra226 (2014).
Garouniatis, A. et al. FAK, CD44v6, c-Met and EGFR in colorectal cancer parameters: tumour progression, metastasis, patient survival and receptor crosstalk. Int J Colorectal Dis 28, 9–18 (2013).
Amirouchene-Angelozzi, N., Swanton, C. & Bardelli, A. Tumor Evolution as a Therapeutic Target. Cancer discovery (2017).
Pietrantonio, F. et al. RET fusions in a small subset of advanced colorectal cancers at risk of being neglected. Annals of oncology: official journal of the European Society for Medical Oncology 29, 1394–1401 (2018).
Pietrantonio, F. et al. O-011RET rearrangements define a new and rare molecular subtype of metastatic colorectal cancer (mCRC). Annals of Oncology 28, mdx262.010–mdx262.010 (2017).
Piotrowska, Z. et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer discovery 8, 1529–1539 (2018).
Vaishnavi, A. et al. EGFR Mediates Responses to Small-Molecule Drugs Targeting Oncogenic Fusion Kinases. Cancer Res 77, 3551–3563 (2017).
Le Rolle, A. F. et al. Identification and characterization of RET fusions in advanced colorectal cancer. Oncotarget 6, 28929–28937 (2015).
Luraghi, P. et al. A molecularly annotated model of patient-derived colon cancer stem-like cells to assess genetic and non-genetic mechanisms of resistance to anti-EGFR therapy. Clin Cancer Res (2017).
Drilon, A. et al. Response to ERBB3-Directed Targeted Therapy in NRG1-Rearranged Cancers. Cancer discovery 8, 686–695 (2018).
Prentice, L. M. et al. NRG1 gene rearrangements in clinical breast cancer: identification of an adjacent novel amplicon associated with poor prognosis. Oncogene 24, 7281–7289 (2005).
CancerGenomeAtlasResearchNetwork. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337 (2012).
Khan, K. H. et al. Abstract 4339: Molecular subtypes and novel genetic mechanisms of primary and acquired anti-EGFR resistance in colorectal cancer in the Prospect C biomarker trial. Cancer Research 78, 4339–4339 (2018).
Guo, C. et al. ZEB1 Promotes Oxaliplatin Resistance through the Induction of Epithelial - Mesenchymal Transition in Colon Cancer Cells. J Cancer 8, 3555–3566 (2017).
Fukuda, K. et al. Epithelial-to-Mesenchymal Transition Is a Mechanism of ALK Inhibitor Resistance in Lung Cancer Independent of ALK Mutation Status. Cancer Res 79, 1658–1670 (2019).
Copps, K. D. & White, M. F. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia 55, 2565–2582 (2012).
Hancer, N. J. et al. Insulin and metabolic stress stimulate multisite serine/threonine phosphorylation of insulin receptor substrate 1 and inhibit tyrosine phosphorylation. J Biol Chem 289, 12467–12484 (2014).
Shah, O. J. & Hunter, T. Turnover of the active fraction of IRS1 involves raptor-mTOR- and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Molecular and cellular biology 26, 6425–6434 (2006).
DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics 43, 491–498 (2011).
Greenman, C. D. et al. PICNIC: an algorithm to predict absolute allelic copy number variation with microarray cancer data. Biostatistics 11, 164–175 (2010).
Van Loo, P. et al. Allele-specific copy number analysis of tumors. Proceedings of the National Academy of Sciences of the United States of America 107, 16910–16915 (2010).
Talevich, E., Shain, A. H., Botton, T. & Bastian, B. C. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS computational biology 12, e1004873 (2016).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nature medicine 21, 1350–1356 (2015).
Lee, E., Chuang, H. Y., Kim, J. W., Ideker, T. & Lee, D. Inferring pathway activity toward precise disease classification. PLoS computational biology 4, e1000217 (2008).
Uhlik, M. T. et al. Stromal-Based Signatures for the Classification of Gastric Cancer. Cancer Res 76, 2573–2586 (2016).
Lee, J. K. et al. Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy. Nature genetics (2018).
Acknowledgements
This work was supported by a grant from the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (HI14C2750 to YSP, HI14C3418 to JL), and the SMC Biobank (to HCK and JL).
Author information
Authors and Affiliations
Contributions
The study concept and design were by S.M.B., J.L., H.C.K. and C.R. J.L., S.T.K., Y.S.P., J.O.P., H.Y.L., W.K.K. and H.C.K. acquired and managed patients, provided facilities, etc. Analysis and interpretation of data were performed by S.M.B., S.T.K., J.U.H., P.J.E., J.N.C., I.S.W., T.G., S.S.W., H.R.Q., J.C.T., J.L., M.D.W., R.D.N., A.A. and C.R. All the co-authors were involved in writing and reviewing the paper. The study was supervised by J.L., H.C.K. and C.R.
Corresponding authors
Ethics declarations
Competing interests
S.B., P.J.E., J.N.C., I.H.W., T.G., S.S.W., H.Q., J.C.T., J.L., M.W., R.D.N., A.A. and C.R. are employees of Lilly. J.L., S.T.K., S.Y.K., J.Y.H., Y.S.P., J.O.P., H.L., W.K.K. and H.C.K. have no conflicts of interest to declare.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bray, S.M., Lee, J., Kim, S.T. et al. Genomic characterization of intrinsic and acquired resistance to cetuximab in colorectal cancer patients. Sci Rep 9, 15365 (2019). https://doi.org/10.1038/s41598-019-51981-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-51981-5
This article is cited by
-
Histological transformation to signet-ring cell carcinoma in a patient with clinically aggressive poorly differentiated adenocarcinoma of the ascending colon after response to chemotherapy plus cetuximab: a case report
World Journal of Surgical Oncology (2023)
-
FGFR1 is a potential therapeutic target in neuroblastoma
Cancer Cell International (2022)
-
Senescence-associated reprogramming induced by interleukin-1 impairs response to EGFR neutralization
Cellular & Molecular Biology Letters (2022)
-
Dynamic increase of M2 macrophages is associated with disease progression of colorectal cancers following cetuximab-based treatment
Scientific Reports (2022)
-
A tumor microenvironment-specific gene expression signature predicts chemotherapy resistance in colorectal cancer patients
npj Precision Oncology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.