Abstract
The molecular pathogenesis of gastroenteropancreatic (GEP) neuroendocrine tumours (NETs) remains to be elucidated. High-mobility group A (HMGA) proteins play important roles in the regulation of transcription, differentiation, and neoplastic transformation. In this study, the expression of HMGA1 and HMGA2 was studied in 55 GEP NETs. Overexpression of HMGA1 and 2 was frequently detected in GEP NETs compared with normal tissues. Nuclear immunostaining of HMGA1 and 2 was observed in GEP NETs (38 of 55, 69%; 40 of 55, 73%, respectively). High-mobility group A2 expression increased from well-differentiated NET (WNET) to well-differentiated neuroendocrine carcinoma (WNEC) and poorly differentiated NEC (PNEC) (P<0.005) and showed the highest level in stage IV tumours (P<0.01). In WNECs, the expression of HMGA1 and 2 was significantly higher in metastatic tumours than those without metastasis (P<0.05). Gastroenteropancreatic NETs in foregut showed the highest level of HMGA1 and 2 expressions. MIB-1 labelling index (MIB-1 LI) correlated with HMGA1 and 2 overexpression (R=0.28, P<0.05; R=0.434, P<0.001; respectively) and progressively increased from WNETs to WNECs and PNECs (P<0.001). Let-7 expression was addressed in 6 normal organs, 30 tumour samples, and 24 tumour margin non-tumour tissues. Compared with normal tissues, let-7 downregulation was frequent in NETs (19 of 30, 63%). Higher expression of HMGA1 and 2 was frequently observed in tumours with let-7 significant reduction (53, 42%, respectively). The reverse correlation could be detected between HMGA1 and let-7 (P<0.05). Our findings suggested that HMGA1 and 2 overexpression and let-7 downregulation might relate to pathogenesis of GEP NETs.
Similar content being viewed by others
Main
Although the gastrointestinal (GI) tract and pancreatic organ have the largest population of endocrine cells, gastroenteropancreatic (GEP) neuroendocrine tumours (NETs) are rare with an age-adjusted annual incidence of 2.5–4.5 per 100 000 (Maggard et al, 2004). However, their incidence is increasing by approximately 6% per year (Gustafsson et al, 2008). Gastroenteropancreatic NETs comprise a heterogeneous group of neoplasms arising from the diffuse neuroendocrine system and are divided by their embryological site of origin into foregut, midgut, and hindgut tumours (Maggard et al, 2004). These tumours strongly differ from each other on the basis of different pathogenetic, clinical, functional, histological, and prognostic patterns (Klöppel et al, 2004). The molecular mechanisms of GEP NETs tumourigenesis are poorly understood (Zikusoka et al, 2005).
The high-mobility group A (HMGA) proteins are non-histone chromosomal proteins that bind through their AT-binding motifs to the minor groove of AT-rich DNA strands. They have no intrinsic transcriptional activity but can modulate transcription by altering chromatin architecture (Reeves and Nissen, 1990; Thanos and Maniatis, 1992; Thanos et al, 1993). The two types of HMGA protein, HMGA1 and HMGA2, have similar functions and are encoded by two different genes at chromosomal loci 6p21.3 and 12q15, respectively (Johnson et al, 1989). High-mobility group A proteins are widely expressed during embryogenesis, whereas their expression is low or absent in normal adult tissues (Zhou et al, 1995; Chiappetta et al, 1996). High-mobility group A proteins are involved in many diverse biological processes such as regulation of transcription, embryogenesis, differentiation, neoplastic transformation, and integration, and expression of viral genomes (Sgarra et al, 2004). Both HMGA1 and 2 proteins are overexpressed in several experimental and human malignancies. Overexpression of HMGA1 has been noted in different cancer types and it has been recognised as a good target for therapeutic intervention in pancreatic cancer. High-mobility group A2 overexpression is a hallmark of various benign and malignant tumours and is also associated with a highly malignant phenotype and is a poor prognostic index (Fusco and Fedele, 2007). However, there is no study to detect HMGA proteins expression and to evaluate their role in GEP NETs.
The microRNAs (miRNA), a family of mature noncoding small RNA 21–25 length nucleotides, have been shown to play a role in a variety of biological processes. They regulate gene expression at the posttranscription level by direct cleavage of a target mRNA using interference machinery (mRNA cleavage) or by inhibition of protein synthesis (Caldas and Brenton, 2005; Esquela-Kerscher and Slack, 2006). Recent evidence has shown that miRNA misexpression correlates with various human cancers and indicated that some miRNAs can function as oncogenes or tumour suppressors (Esquela-Kerscher and Slack, 2006). Several studies reported that HMGA2 is negatively regulated by the let-7 miRNA family in a mouse model system (Mayr et al, 2007), in head and neck cancers (Hebert et al, 2007), in uterine leiomyomas (Wang et al, 2007), in lung cancer cell lines (Lee and Dutta, 2007), and in ovarian cancers (Shell et al, 2007). By computer searches (TargetScan, TargetScanS, PicTar, and miRNAviewer), HMGA1 can also be potentially regulated by let-7. To our knowledge, there are few reports about miRNA expression in GEP NETs and no investigation about the relations between HMGA1 and 2 proteins expression and the let-7 expression in GEP NETs.
In this study, we investigated the prevalence of HMGA1 and 2 involving in different GEP NETs and assessed its relevance to malignancy. We performed a systematic analysis of immunohistochemistry and reverse transcription (RT)–PCR for HMGA1 and 2 expression in a series of GEP NETs from different sites. Let-7 was also analysed by semiquantitative RT–PCR. The data were then correlated with relevant clinical information. In addition, we investigated possible associations between HMGA1, 2 expression and let-7 expression.
Materials and methods
Patients and samples
Fifty-five paraffin-embedded surgical or biopsy specimens were collected (from the surgical pathology files of the Department of Pathology, University of Tokushima School of Medicine, and the affiliated hospitals) from patients affected by NETs of GEP from 1982 to 2007. Tumours of the GEP tract were from foregut: esophagous (n=1), stomach (n=11), pancreas (n=11), duodenum (n=6); midgut: ileum (n=2), appendix (n=1); hindgut: colon (n=5), rectum (n=14); liver metastasis (n=1); and lymph node metastasis (n=3). The frequency of GEP NETs in this study is general agreement with published reports from the Neuroendocrine Tumour Workshop of Japan (Ito et al, 2007). Notably, the frequency of midgut NETs was very low. Paraffin sections were stained with pan-NE markers, such as chromogranin A and synaptophysin, to confirm the NE immunophenotype. The diagnosis of NETs was made on the basis of morphological and immunohistochemical findings evaluated by two independent pathologists (ZRQ and TS), according to the World Health Organization classification (Klöppel et al, 2004). These included well-differentiated NET (WNET, of either benign or uncertain behaviour; n=29), well-differentiated NEC (WNEC; n=13), and poorly differentiated NEC (PNEC; n=13). The tumour stage was made on the basis of the recently published TNM classification system (Rindi et al, 2006; Rindi et al, 2007). Patient characteristics for these cases are summarised in Table 1.
Immunohistochemistry
High-mobility group A1, HMGA2, and Ki-67 antigen immunolocalisation based on the labelled streptavidin-biotin method were performed on sections from representative blocks of paraffin-embedded tissues. After deparaffinisation and antigen retrieval using an autoclave oven technique, sections were incubated at 4°C overnight with goat polyclonal anti-HMGA1 antibody (1 : 50; Santa Cruz Biotech, Santa Cruz, CA, USA) and goat polyclonal anti-HMGA2 antibody (1 : 50; Santa Cruz Biotech) or with Ki-67 antigen mouse monoclonal antibody (1 : 100, DakoCytomatin, Glostrup, Denmark). Antigen–antibody complexes were detected using the cobalt-3, 3′-diaminobenzidine (Co-DAB) reaction. Squamous cell carcinomas known to be positive for HMGA1 and 2 were used as positive control. Sections incubated in PBS without the primary antibody served as negative controls. Owing to the absence of tissue entirely composed of neuroendocrine cells, several whole normal human organs were used as controls: oesophagus, stomach, duodenum, appendix, small intestine, pancreas, colon, and rectum. Clear nuclear staining was considered to present a positive stain for HMGA1, 2, and Ki-67. A total of 200–1000 cells were counted and the percentage of HMGA1-, 2-stained tumour cells was scored on a scale of 0–3 (0: no expression; 1+: 1–10%; 2+: 10–50%; 3+: >50%). The Ki-67 antigen labelling index (LI) was determined by counting the number of positive cells in a total of 200–1000 tumour cells observed in several representative high-power fields ( × 400).
Microdissection and RNA isolation
From each paraffin block of representative tumour areas, serial sections with a thickness of 10 μm were prepared and stained with nuclear Fast Red (Sigma-Aldrich, Tokyo, Japan). Malignant cells were selected under microscope magnification ( × 20 to × 100) and dissected from the slide using a scalpel or Laser microdissection (Molecular Machines & Industries, Glattbrugg, Switzerland). Total RNA was isolated using the high pure RNA paraffin kit (Roche Diagnostics GmbH, Mannheim, Germany) as per the manufacturer's instructions. Extracted RNA concentration and purity were measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Del, Wilmington, DE, USA) and by PCR using the primer pair specific for GAPDH with or without RT, followed by 1% agarose gel electrophoresis and ethidium bromide staining.
RT–PCR analysis for HMGA1 and 2
Total RNA (500 ng) was reverse transcribed, using random hexanucleotides as primers (50 pM) and 200 units primescript reverse transcriptase (Takara Bio Inc., Otu, Japan). The cDNA was amplified in a 20 μl reaction mixture containing 0.2 mM dNTP, 1.0 mM MgSO4, 0.5 μ M each primer and 1 U KOD-Plus polymerase (Toyobo, Osaka, Japan). After a denaturing step (95°C for 2 min), the cDNA was further amplified in 35 PCR cycles (95°C for 1 min, 58°C for 30 s, and 68°C for 30 s). The following primers were used to amplify the HMGA1 transcript: forward primer, 5-AGAGACCTCGGGGCCGACCA-3; reverse primer, 5-GATGCCCTCCTCTTCCTCCTT-3. The following primers were used to amplify the HMGA2 transcript: forward primer, 5-ACTTCAGCCCAGGGACAAC-3, which maps onto the first exon; reverse primer, 5-GCTGCTTTAGAGGGACTCTTGTT-3, which maps onto the second exon. Expression of the GAPDH gene was used as an internal control for the amount of cDNA tested. The reaction products were analysed on a 2% agarose gel. Total RNA samples from several whole normal human organs were used as controls: oesophagus, stomach, duodenum, appendix, small intestine, pancreas, colon, and rectum.
Semiquantitative RT–PCR analysis for let-7
The expression of human mature let-7 (hsa-let-7) miRNA was analysed using a mirVana quantitative RT–PCR miRNA detection kit (Ambion, Austin, TX, USA, Cat. nos. AM1558 and AM30000) according to the manufacturer's protocol. Ubiquitously expressed U6 small nuclear RNA (Ambion, Cat. No. AM30303) was used for normalisation and as an internal control. Briefly, RT–PCR was performed with 20 ng of total RNA using gene-specific RT primers. cDNA was generated. The PCR consisted of 32 cycles (95°C for 15 s, 60°C for 30 s) after an initial denaturation step (95°C for 3 min). The PCR products were analysed by electrophoresis on 2% agarose gels. Quantitation of let-7 expression levels was achieved by densitometric scanning of ethidium bromide-stained gels. The levels of expression of let-7 was analysed with computer software (Image, NIH) and shown as the ration of let-7 to U6 (Qian et al, 2005).
Statistical analysis
To determine the significance of associations between different variables, data were statistically analysed by Mann–Whitney U-test, Kruskal–Wallis test, χ2–test, and Spearman's correlation coefficient using StatView J-4.5 software (Abacus Concepts, Berkeley, CA, USA). A P-value of less than 0.05 was considered statistically significant.
Results
HMGA1 and 2 expression analysis by IHC
Immunolocalisation of HMGA1, 2 proteins was mainly nuclear. However, cytoplasmic staining was observed similar to that reported earlier for placental extravillous trophoblast and lung cancer, but was found to be nonspecific background staining and was ignored (Sarhadi et al, 2006).
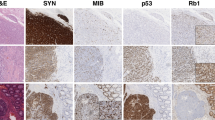
In general, there were no HMGA1 and 2 expressions in the normal organs including oesophagus, stomach, duodenum, appendix, small intestine, pancreas, colon, and rectum. In GEP normal NE cells, detected by choromgranin A (Figure 1A–E), HMGA1 and 2 were immunostained negatively (Figure 1F–O).
In GEP NETs, a high proportion of tumours (38 of 55, 69%) was detected with nuclear HMGA1 protein expression (Figure 2E–H, Tables 1 and 2). Twenty-seven (49%) tumours showed a high expression (3+ and 2+), 13 (24%) tumours showed a low expression (1+), and 17 (31%) tumours were immuno negative. According to different histopathologic categories, HMGA1 overexpression was detected in 21/29 (72%) of WNETs, 8/13 (61%) of WNECs and 9/13 (70%) of PNECs (Table 2). This HMGA1 protein expression did not show a significant difference level among WNETs, WNECs and PNECs (Figure 3A). However, NETs in foregut showed the highest level of HMGA1 expression even it was not significant (Figure 3C).
Haematoxilin and eosin staining, high-mobility group A1 (HMGA1) and HMGA2 immunostaining figures of well-differentiated NET (WNET) (A, E and I), insulinoma (B, F, and J), well-differentiated neuroendocrine carcinoma (WNEC) (C, G, and K), and poorly differentiated NEC (PNEC) (D, H, and L), respectively. High-mobility group A1 (E–H) and HMGA2 (I–L) showed strongly nuclear immunostaining in gastroenteropancreatic neuroendocrine tumours.
The relations between expression levels of high-mobility group A (HMGA1), HMGA2 proteins and clinicopathologic data of gastroenteropancreatic (GEP) neuroendocrine tumours (NETs). According to different histopathologic categories, HMGA1 protein expression did not show significant difference among well-differentiated NET (WNET), well-differentiated neuroendocrine carcinoma (WNEC), and poorly differentiated NEC (PNEC) (A). High-mobility group A2 protein expression increased from WNET to WNEC and PNEC, and its significant difference was observed between WNET and PNEC (P<0.005) (B). Gastroenteropancreatic NETs in foregut showed the highest level of HMGA1 expression but it was not significant (C). Gastroenteropancreatic NETs of foregut also showed the highest level of HMGA2 expression (P<0.05) (D). In GEP WNECs, the expression of HMGA1 and 2 was higher in metastasis tumours than in tumours without metastasis (P<0.05, P<0.05) (E and F). High-mobility group A1 protein expression increased following tumour stage but did not show significant difference (G). High-mobility group A2 protein showed the highest level in stage IV GEP NETs (P<0.01) (H). MIB LI progressively increased following HMGA1 and 2 expression score, although it was not significant (I and J). MIB-1 LI progressively increased from WNETs to WNECs and PNECs (P<0.001) (K), and following tumour stage (P<0.05) (L).
A clear nuclear staining for HMGA2 was detected in 40 of 55 (73%) GEP NETs (Figure 2I–L, Tables 1 and 2). Twenty-five of these (46%) showed a high expression (3+ and 2+) of HMGA2, and 15 (27%) tumours showed low expression (1+), and 15 (27%) tumours were negative immunoreactions (Table 2). According to different histopathologic categories, HMGA2 protein expression progressively increased from WNET to WNEC and PNEC, and the significant increasing was detected between WNET and PNEC (P<0.005) (Figure 3B). Neuroendocrine tumours of foregut showed the frequent HMGA2 overexpression and highest level of HMGA2 expression (P<0.05) (Table 2, Figure 3D).
In WNECs, the expression of HMGA1 and 2 was significantly higher in metastatic tumours than in tumours without metastasis (P<0.05, P<0.05; respectively; Figure 3E and F). This tendency is not clear in PNECs. In addition, HMGA2 showed the highest level in stage IV GEP NETs (stage IV vs stages I, II, III, P<0.01; Figure 3H). High-mobility group A1 expression was also higher in stage IV tumours than stages I, II, and III tumours, but it was not significant (Figure 3G).
We also investigated the correlation between MIB-1 labelling index (MIB-1 LI), which was assessed earlier to enroll patients in the clinical trial and HMGA proteins expression. High-mobility group A1 and 2 nuclear staining positively correlated with the proliferation index in GEP NETs (R=0.28, P<0.05; R=0.434, P<0.001; respectively). MIB-1 LI progressively increased following HMGA1 and 2 expression score, although not significant (Figure 3I and J). MIB-1 LI also progressively increased from WNETs to WNECs and PNECs (P<0.001; Figure 3K) and following tumour stage (P<0.05; Figure 3L). Furthermore, MIB-1 LI positively correlated with tumour size (R=0.409, P<0.005).
Between 6 GI WNETs with uncertain behaviour (>1 cm or vascular invasion) and 14 GI WNETs with benign behaviour, HMGA1 and 2 expressions did not show any significant differences (data not shown). Although HMGA1 and 2 expressions were potentially high in WNECs, there was no significant correlation between HMGA proteins expression and tumour malignancy (i.e., WNETs vs WNECs) in foregut or hindgut NETs, respectively (data not shown). We could not find any significant differences in HMGA1 and 2 expression between PNEC areas and adenocarcinoma areas in several PNEC cases (data not shown).
HMGA1 and 2 mRNA expression analysis by RT–PCR
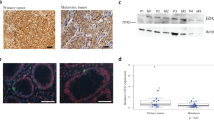
In RT–PCR mRNA study, HMGA1 and 2 mRNA expression was analysed on normal tissues from 7 organs (each organ sample pool included RNA from four normal tissues) and 31 tumours with 25 surrounding non-tumour tissues. As shown in Figure 4C, HMGA1 and 2 mRNA amplification was not observed in normal organs. Only a very weak band could be seen in colon and rectum. However, in many tumours HMGA1 and 2 mRNA was abundantly amplified, whereas their expression was not detected in surrounding non-tumour tissues (Table 1, Figure 4D and E). Generally, the mRNA expression of HMGA1and 2 correlated well with the nuclear expression of HMGA1 and 2 proteins as detected by IHC analysis (P<0.0005, P<0.0005, respectively).
Pre-microdissection figure of one well-differentiated neuroendocrine tumour (WNET) (A). Post-microdissection figure of the same WNET (B). High-mobility group A1 (HMGA1) (133 bp) and HMGA2 (155 bp) mRNA amplification were not observed in normal tissues. In colon and rectum, a very weak band was detected (C). But in tumours, HMGA1 (D) and 2 (E) mRNA was abundantly amplified, whereas their expressions were not detected in tumour margin non-tumour tissues. GAPDH (375 bp) expression was used as endogenous control for RNA integrity.
Expression of let-7 in GEP NETs
Semiquantitative RT–PCR method was used to address the levels of let-7 in normal tissues from six organs including stomach, pancreas, duodenum, small intestine, appendix, and colon (each organ sample pool included RNA from four normal tissues). Normal pituitary, used as positive control, expressed easily detectable levels of let-7 (let-7/U6, 0.9) (Figure 5A). Other normal samples from six organs showed moderate levels of let-7 (let7/U6, 0.21–0.24) (Figure 5A). This difference may relate to tissue specificity. In tumour margin tissue samples, 22 of 24 cases expressed high levels of let-7 (let-7/U6, 0.33–1.49) (Table 1, Figure 5B). On the basis of the above observation, we arbitrarily classified expression levels of less than one-half of let-7 value in normal tissues, that is, let-7/U6 <0.1 as significant reduction. Thus, downregulated expression of let-7 was found in 19 of 30 (63%) tumours (negative, 11; significantly reduction, 9). The average levels of let-7 expression were significantly lower in GEP NETs than normal organs and their surrounding non-tumour tissues (Figure 5C). Let-7 expression was not significantly different among WNETs, WNECs, and PNECs (Figure 5C, Table 3).
Semiquantitative reverse transcription–PCR method was used to address the levels of let-7 in normal tissues from 6 organs (stomach, pancreas, duodenum, small intestine, appendix, and colon), 30 gastroenteropancreatic neuroendocrine tumours (NETs), and their margin non-tumour tissues. Normal pituitary tissue was used as positive control. U6 expression was used as endogenous control for small RNA integrity and as templates for quantitative analysis. Generally, normal tissues from different organs clearly showed let-7 expression (A). Comparing with the level of let-7 expression in tumour's margin non-tumour tissues, negative or reduced expression of let-7 was detected in tumours (B). The average level of let-7 expression was significantly lower in well-differentiated NET (WNETs), well-differentiated NECs (WNECs), and poorly differentiated NEC (PNECs) than in their margin non-tumour tissues and in normal organs (*P<0.05). Interestingly, the let-7 expression was higher in tumour's margin non-tumour tissues than in normal organs. In addition, let-7 expression was not significantly different among WNETs, WNECs, and PNECs (C).
We also investigated the relationship between let-7 expression and HMGA1, 2 proteins expression in 30 GEP NETs through several viewpoints. High-mobility group A1 high expression was more frequently detected in 19 tumours with significant let-7 downregulation (53%) than in tumours with regular level of let-7 expression (P<0.05). The average level of let-7 expression was lower in NETs with HMGA1 overexpression than in tumours with negative or moderate expression of HMGA1 (data not shown). These data implied that there might be inverse correlation between the expression of let-7 and HMGA1. Unfortunately, we could not observe the clear inverse correlation between the expression of let-7 and HMGA2 protein expression, although high expression of HMGA2 was frequently detected in tumours with significant downregulation of let-7 (42%). When analysed the relation between clinicopathological characteristics of 30 GEP NETs and reduced let-7 expression, there were no notable differences in patient age, sex, tumour size, vascular invasion, or tumour metastasis (data not shown).
Discussion
Gastroenteropancreatic NETs originate from the cells of the diffuse endocrine system. The molecular genetic mechanism of development and progression is complex and remains largely unknown. Gastroenteropancreatic NETs do not show alterations in oncogenes, such as ras, myc, fos jun, and src, or in common tumour suppressor genes (Delle Fave and Corleto, 2001). High-mobility group A1 and 2 have been detected in many kinds of benign and malignant tumours (Fusco and Fedele, 2007). Interestingly, overexpression of HMGA1 and 2 also has been shown in NETs including small cell carcinomas and pituitary adenomas (Brenner et al, 2004; Fusco and Fedele, 2007). These findings enhanced us to investigate the role of HMGA1 and 2 protein in GEP NETs tumourigenesis and progression.
Our study is the first time to characterise HMGA1 and 2 overexpression in GEP NETs. We found that HMGA1 and 2 overexpression was a common event in GEP NETs. Notably, a high level overexpression of HMGA1 and 2 was frequently observed in WNETs as in WNECs and PNECs. High-mobility group A proteins contributed to tumourigenesis in various benign human tumours including lipomas, uterine leiomyomas, and pituitary adenomas. Little is known about pathogenesis of sporadic WNETs (Zikusoka et al, 2005). Our findings suggested that HMGA proteins might relate to the tumourigenesis of GEP NETs.
High-mobility group A1 and 2 interact with many different transcription factors and influence numerous gene expression patterns. They are important regulators of cell growth, differentiation, apoptosis, and transformation (Reeves, 2001). It has been shown that a high expression of HMGA1 and 2 is associated with a highly malignant phenotype and is a poor prognostic index (Fusco and Fedele, 2007). The expression of HMGA1 differed from the expression of HMGA2 in GEP NETs only with respect to the correlation of expression with histopathologic categories. High-mobility group A2 protein expression increased from WNETs to WNECs and PNECs and from stages I, II, III tumours to stage IV tumours. Interestingly, both HMGA1 and 2 overexpression may relate to tumour metastasis in WNECs. Furthermore, we observed the correlation between cell proliferation marker MIB-1 LI and HMGA protein overexpression. Because more than a half of GEP NETs are functionally inactive and are usually diagnosed once signs and symptoms of tumour metastasis occur (Oberg, 2002), the distinction benign and malignant GEP NET is very important. However, it has not been completely resolved (Rindi and Bordi, 2005). Our data implied that HMGA proteins may reliably predict the course of this disease and may have prognostic significances in GEP NETs. However, the exact mechanism involving in HMGA oncogenic activity in GEP NET needs further investigation.
The current literature suggests that several genes are involved in GEP NET tumourigenesis with significant differences among tumours of different embryological derivatives: foregut, midgut, and hindgut. The MEN1 gene is involved in initiation of 33% of foregut GEP NETs (Lubensky and Zhuang, 2007). 18q defects are present almost exclusively in mid/hindgut NETs (Lubensky and Zhuang, 2007). X-chromosome markers are associated with malignant behaviour in foregut tumours only (Lubensky and Zhuang, 2007). In this study, GEP NETs in foregut showed the highest level of HMGA1 and 2 proteins expression. However, the differences in HMGA protein expression among tumour sites need further investigation in large series of GEP NETs.
Recent studies have shown that let-7 plays a role as tumour suppressors by negatively regulating expression of RAS and HMGA2 oncogenes (Johnson et al, 2005; Hebert et al, 2007; Mayr et al, 2007; Wang et al, 2007). Let-7 downregulation is commonly seen in neoplasm, including lung, breast and gastric cancers, and uterine leiomyomas (Takamizawa et al, 2004; Iorio et al, 2005; Wang et al, 2007; Motoyama et al, 2008). In this study, we observed that let-7 downregulation is very common from benign NETs to small cell carcinomas. This is the first finding that aberrant expression of let-7 may relate to GEP NET tumourigenesis. Furthermore, the reverse correlation between let-7 downregulation and HMGA1 overexpression has been first observed in tumours. Till now, the mechanism of HMGA1 overexpression is not clear and many studies only focus on HMGA2 regulated by let-7 family (Takamizawa et al, 2004; Iorio et al, 2005; Wang et al, 2007; Motoyama et al, 2008). Our findings implied that loss or reduction of let-7 might be one potential mechanism of HMGA1 protein overexpression. On the other hand, although the clearly reverse correlation between let-7 downregulation and HMGA2 overexpression was not detected in this small series of GEP NETs, loss and reduction of let-7 expression may also be one important mechanism of HMGA2 overexpression. The truncated transcripts of HMGA2 with a partial or complete loss of let-7 complementary sites can explain the increased expression of HMGA2 in tumours with slight reduction or regular level of let-7 expression (Fusco and Fedele, 2007). In addition, by computer searches, we found that HMGA1 and 2 can be potentially regulated by many other miRNAs. Further investigation of miRNAs would be important in GEP NET tumourigenesis study.
Several studies have aimed to develop cancer therapy by inhibiting HMGA proteins (Scala et al, 2000). In pancreatic adenocarcinoma, blocking HMGA protein synthesis has a negative effect on tumour cell proliferation and metastatic potential (Liau et al, 2006). The differential expression of HMGA1 and 2 in GEP NETs and normal cells should allow for the specificity and lower toxicity of such therapy. Furthermore, as HMGA1 and 2 proteins are frequently overexpressed in GEP NETs, HMGA-targeted anticancer therapy could have a wide ranged application.
Change history
16 November 2011
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Brenner B, Tang LH, Klimstra DS, Kelsen DP (2004) Small-cell carcinomas of the gastrointestinal tract: a review. J Clin Oncol 22: 2730–2739
Caldas C, Brenton JD (2005) Sizing up miRNAs as cancer genes. Nat Med 11: 712–714
Chiappetta G, Avantaggiato V, Visconti R, Fedele M, Battista S, Trapasso F, Merciai BM, Fidanza V, Giancotti V, Santoro M, Simeone A, Fusco A (1996) High level expression of the HMGI (Y) gene during embryonic development. Oncogene 13: 2439–2446
Delle Fave G, Corleto VD (2001) Oncogenes, growth factors, receptor expression and proliferation markers in digestive neuroendocrine tumours. A critical reappraisal. Ann Oncol 2S: 13–17
Esquela-Kerscher A, Slack FJ (2006) Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269
Fusco A, Fedele M (2007) Roles of HMGA proteins in cancer. Nat Rev Cancer 7: 899–910
Gustafsson BI, Kidd M, Modlin IM (2008) Neuroendocrine tumours of the diffuse neuroendocrine system. Curr Opin Oncol 20: 1–12
Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ (2007) High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer 6: 5
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65: 7065–7070
Ito T, Tanaka M, Sasano H, Osamura YR, Sasaki I, Kimura W, Takano K, Obara T, Ishibashi M, Nakao K, Doi R, Shimatsu A, Nishida T, Komoto I, Hirata Y, Imamura M, Kawabe K, Nakamura K, Neuroendocrine Tumour Workshop of Japan (2007) Preliminary results of a Japanese nationwide survey of neuroendocrine gastrointestinal tumours. J Gastroenterol 42: 497–500
Johnson KR, Lehn DA, Reeves R (1989) Alternative processing of mRNAs encoding mammalian chromosomal high-mobility-group proteins HMG-I and HMG-Y. Mol Cell Biol 9: 2114–2123
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120: 635–647
Klöppel G, Perren A, Heitz PU (2004) The gastroenteropancreatic neuroendocrine cell system and its tumours: the WHO classification. Ann N Y Acad Sci 1014: 13–27
Lee YS, Dutta A (2007) The tumour suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21: 1025–1030
Liau SS, Jazag A, Whang EE (2006) HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res 66: 11613–11622
Lubensky IA, Zhuang Z (2007) Molecular genetic events in gastrointestinal and pancreatic neuroendocrine tumours. Endocr Pethol 18: 156–162
Maggard MA, O’Connell JB, Ko CY (2004) Updated population-based review of carcinoid tumours. Ann Surg 240: 117–122
Mayr C, Hemann MT, Bartel DP (2007) Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315: 1576–1579
Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M (2008) Clinical significance of high mobility group A2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res 14: 2334–2340
Oberg K (2002) Carcinoid tumours: molecular genetics, tumour biology, and update of diagnosis and treatment. Curr Opin Oncol 14: 38–45
Qian ZR, Sano T, Yoshimoto K, Yamada S, Ishizuka A, Mizusawa N, Horiguchi H, Hirokawa M, Asa SL (2005) Inactivation of RASSF1A tumour suppressor gene by aberrant promoter hypermethylation in human pituitary adenomas. Lab Invest 85: 464–743
Reeves R (2001) Molecular biology of HMGA proteins: hubs of nuclear function. Gene 277: 63–81
Reeves R, Nissen MS (1990) The AT DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem 265: 8573–8582
Rindi G, Bordi C (2005) Endocrine tumours of the gastrointestinal tract: aetiology, molecular pathogenesis and genetics. Best Pract Res Clin Gastroenterol 19: 519–534
Rindi G, Klöppel G, Alhman H, Caplin M, Couvelard A, de Herder WW, Erikssson B, Falchetti A, Falconi M, Komminoth P, Körner M, Lopes JM, McNicol AM, Nilsson O, Perren A, Scarpa A, Scoazec JY, Wiedenmann B, all other Frascati Consensus Conference participants; European Neuroendocrine Tumoursociety (ENETS) (2006) TNM staging of foregut (neuro)endocrine tumours: a consensus proposal including a grading system. Virchows Arch 449: 395–401
Rindi G, Klöppel G, Couvelard A, Komminoth P, Körner M, Lopes JM, McNicol AM, Nilsson O, Perren A, Scarpa A, Scoazec JY, Wiedenmann B (2007) TNM staging of midgut and hindgut (neuro) endocrine tumours: a consensus proposal including a grading system. Virchows Arch 451: 757–762
Sarhadi VK, Wikman H, Salmenkivi K, Kuosma E, Sioris T, Salo J, Karjalainen A, Knuutila S, Anttila S (2006) Increased expression of high mobility group A proteins in lung cancer. J Pathol 209: 206–212
Scala S, Portella G, Fedele M, Chiappetta G, Fusco A (2000) Adenovirus-mediated suppression of HMGI(Y) protein synthesis as potential therapy of human malignant neoplasias. Proc Natl Acad Sci USA 97: 4256–4261
Sgarra R, Rustighi A, Tessari MA, Di Bernardo J, Altamura S, Fusco A, Manfioletti G, Giancotti V (2004) Nuclear phosphoproteins HMGA and their relationship with chromatin structure and cancer. FEBS Lett 574: 1–8
Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME (2007) Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci USA 104: 11400–11405
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64: 3753–3756
Thanos D, Du W, Maniatis T (1993) The high mobility group protein HMG I(Y) is an essential structural component of a virus-inducible enhancer complex. Cold Spring Harb Symp Quant Biol 58: 73–81
Thanos D, Maniatis T (1992) The high mobility group protein HMG I(Y) is required for NF-κB-dependent virus induction of the human IFN-β gene. Cell 27: 777–789
Wang T, Zhang X, Obijuru L, Laser J, Aris V, Lee P, Mittal K, Soteropoulos P, Wei JJ (2007) A micro-RNA signature associated with race, tumoursize, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer 46: 336–347
Zhou X, Benson KF, Ashar HR, Chada K (1995) Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376: 771–774
Zikusoka MN, Kidd M, Eick G, Latich I, Modlin IM (2005) The molecular genetics of gastroenteropancreatic neuroendocrine tumours. Cancer 104: 2292–2309
Acknowledgements
We declare that there was no any specific grant for this project and no conflict of interest that could be perceived as prejudicing the impartiality of the research reported. We thank professor Saburo Sone and Department of internal medicine and molecular therapeutics for their excellent technical assistance in microdissection experiment. We also thank Ms Noriko Amo and Mr Akihito Amo for their excellent technical assistance.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Rahman, M., Qian, Z., Wang, E. et al. Frequent overexpression of HMGA1 and 2 in gastroenteropancreatic neuroendocrine tumours and its relationship to let-7 downregulation. Br J Cancer 100, 501–510 (2009). https://doi.org/10.1038/sj.bjc.6604883
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.bjc.6604883
Keywords
This article is cited by
-
HMGA1-TRIP13 axis promotes stemness and epithelial mesenchymal transition of perihilar cholangiocarcinoma in a positive feedback loop dependent on c-Myc
Journal of Experimental & Clinical Cancer Research (2021)
-
Prognostic significance of high mobility group A2 (HMGA2) in pancreatic ductal adenocarcinoma: malignant functions of cytoplasmic HMGA2 expression
Journal of Cancer Research and Clinical Oncology (2021)
-
Overexpression of HMGA2 promotes tongue cancer metastasis through EMT pathway
Journal of Translational Medicine (2016)
-
HMGA1 and HMGA2 expression and comparative analyses of HMGA2, Lin28and let-7 miRNAs in oral squamous cell carcinoma
BMC Cancer (2014)
-
Role of microRNA let-7 and effect to HMGA2 in esophageal squamous cell carcinoma
Molecular Biology Reports (2012)