Abstract
Recent studies revealed that a substantial proportion of patients with high-risk B-cell precursor acute lymphoblastic leukemia (BCP-ALL) harbor fusions involving tyrosine kinase and cytokine receptors, such as ABL1, PDGFRB, JAK2 and CRLF2, which are targeted by tyrosine kinase inhibitors (TKIs). In the present study, transcriptome analysis or multiplex reverse transcriptase–PCR analysis of 373 BCP-ALL patients without recurrent genetic abnormalities identified 29 patients with kinase fusions. Clinically, male predominance (male/female: 22/7), older age at onset (mean age at onset: 8.8 years) and a high white blood cell count at diagnosis (mean: 94 200/μl) reflected the predominance of National Cancer Institute high-risk (NCI-HR) patients (NCI-standard risk/HR: 8/21). Genetic analysis identified three patients with ABL1 rearrangements, eight with PDGFRB rearrangements, two with JAK2 rearrangements, three with IgH-EPOR and one with NCOR1-LYN. Of the 14 patients with CRLF2 rearrangements, two harbored IgH-EPOR and PDGFRB rearrangements. IKZF1 deletion was present in 16 of the 22 patients. The 5-year event-free and overall survival rates were 48.6±9.7% and 73.5±8.6%, respectively. The outcome was not satisfactory without sophisticated minimal residual disease-based stratification. Furthermore, the efficacy of TKIs combined with conventional chemotherapy without allogeneic hematopoietic stem cell transplantation in this cohort should be determined.
Similar content being viewed by others
Introduction
Modern therapeutic strategy of pediatric acute lymphoblastic leukemia (ALL), such as minimal residual disease-based risk-adopted treatment, progress of chemotherapy, supportive care and allogeneic hematopoietic stem cell transplantation (allo-HSCT) greatly improved the prognosis of pediatric ALL.1 However, approximately 15–20% patients experience relapse and show dismal prognosis.1 Additionally, late effect is also the considerable problem for the heavily treated patients, especially for the patients who received allo-HSCT. Thus it is quite important to treat high-risk ALL patients such as Ph+ ALL patients with less toxic chemotherapeutic regimen.2
Recent comprehensive genomic analyses revealed the genetic landscape of high-risk pediatric B-cell precursor ALL (BCP-ALL).3, 4, 5, 6, 7 In particular, a number of chimeric fusions, including those involving tyrosine kinase and cytokine receptors, were identified in a subgroup of BCP-ALL designated as Ph-/BCR-ABL-like ALL.6, 7 Because some of these genetic alterations may be treated by molecular targeted therapies, these patients may benefit from specific kinase inhibitor treatment.8, 9 However, a comprehensive analysis of the clinical characteristics of patients with these kinase fusions is necessary, despite recent reports providing clinical information on these patients.7, 10 The present study aimed to clarify the clinical characteristics of Philadelphia-negative BCP-ALL patients with kinase fusions in Japan by performing a genetic analysis of pediatric BCP-ALL to identify patients with kinase fusions. Clinical information was collected retrospectively and data were analyzed to identify prognostic factors in these patients.
materials and Methods
Patient cohort and samples
Diagnostic samples of bone marrow or peripheral blood were obtained from patients with pediatric BCP-ALL enrolled in and treated under clinical trial protocols of the Japan Association of Childhood Leukemia Study Group (JACLS)-ALL02 study (n=1252),11, 12 Tokyo Children’s Cancer Study Group (TCCSG)-L04-16 (n=150), L06-16 (n=194), L07-16 (n=274) and L09-16 (n=607) studies,13 Childhood Cancer and Leukemia Study Group (CCLSG)-ALL2004 study (n=325)14 and Kyushu–Yamaguchi Childhood Cancer Study Group (KYCCSG)-ALL02 study (n=156).15 The diagnosis of BCP-ALL was based on morphological findings of bone marrow aspirates and immunophenotypic analyses of leukemic cells by flow cytometry. Conventional cytogenetic analyses and molecular studies were performed as part of the routine work-up in each protocol.11, 12, 13, 14, 15 Patients with recurrent fusion transcripts, including ETV6-RUNX1, E2A-PBX1, MLL-related fusion and BCR-ABL, and those with high hyperdiploidy were excluded that defined as B-others ALL. Down syndrome-related BCP-ALL was also excluded from this study. Informed consent was obtained from the guardians of patients according to the Declaration of Helsinki, and genetic study protocols were approved by the institutional review boards of the participating institutes.
mRNA sequencing (mRNA-seq) and multiplex reverse transcriptase–PCR (mRT-PCR)
mRNA-seq or mRT-PCR analyses were performed in 373 BCP-ALL patients with sufficient RNA samples. mRNA-seq was performed in 92 patients in the TCCSG cohort and 17 patients in the JACLS cohort (total 109 patients) according to previously described methods.16 Briefly, the cDNA libraries were loaded on to the cBot (Illumina, Inc., San Diego, CA, USA) for clustering on a flow cell and then sequenced using a HiSeq1000 (Illumina). A paired-end run was performed using the SBS Kit (Illumina). Real-time analysis and basecalling was performed using the HiSeq Control Software Version 1.5 (Illumina). The chimeric transcripts were investigated by employing defuse and TopHat-Fusion, algorithms for gene fusion.17 All kinase fusions determined by mRNA-seq were validated by RT-PCR and Sanger sequencing. mRT-PCR was used in 264 patients (68 in the TCCSG cohort, 95 in the JACLS cohort, 83 in the CCLSG cohort and 18 in the KYCCSG cohort) to determine the presence of 15 kinase fusions, including ZMIZ1-ABL1, SNX2-ABL1, FOXP1-ABL1, SFPO-ABL1, EML1-ABL1, NUP214-ABL1, RCSD1-ABL1, ETV6-ABL1, RANBP2-ABL1, STRN3-JAK2, BCR-JAK2, PAX5-JAK2, EBF1-PDGFRB, ATF7IP-PDGFRB and P2RY8-CRLF2.6, 16, 18 The primers used in this study are listed in Supplementary Table S1.6 The consort diagram of the genetic analysis is described in Figure 1. The comparison of clinical characteristics of analyzed and non-analyzed patients in each cohort, such as TCCSG, JACLS, CCLSG and KYCCSG, are summarized in Supplementary Table S2. Although clinical characteristics were not significantly different between the analyzed and non-analyzed groups in TCCSG, CCLSG and KYCCSG cohorts, National Cancer Institute high-risk (NCI-HR) patients were more in the analyzed group in the JACLS cohort.
A consort diagram of genetic analysis of 373 patients. Samples were obtained from patients treated in the TCCSG (n=160), JACLS (n=112), CCLSG (n=83) and KYCCSG (n=18) cohorts. Ninety-two of the 160 in the TCCSG cohort and 17 of the 112 in the JACLS cohort were analyzed by mRNA-seq. The remaining 264 patients were analyzed by mRT-PCR. The kinase-activating fusions were identified in 16 of the 109 by mRNA-seq and in 13 of the 264 by mRT-PCR.
Determination of IKZF1 deletion and JAK2 mutation
To identify the copy number abnormality of IKZF1, PAX5, EBF1, ETV6, CDKN2A, CDKN2B, RB1 and BTG1 in patients with kinase fusions, the SALSA Multiplex Ligation-dependent Probe Amplification (MLPA) Kit P335-A4 (MRC Holland, Amsterdam, The Netherlands) was used as described previously.19 Screening of JAK2 exons 16, 20 and 21 (gene accession number NM 004972) mutations was performed in patients with CRLF2 rearrangement, as described previously.19
Gene set enrichment analysis
Gene expression profiles of the patients’ samples analyzed by mRNA-seq were obtained as previously described.16 To assess similarity of gene expression profile between the kinase fusion-positive cases and the signature of BCR-ABL1-positive cases, gene set enrichment analysis was performed as previously described. 16
Statistical analysis
Event-free survival (EFS) and overall survival (OS) rates were estimated using the Kaplan–Meier method, and differences were compared using the log-rank test. A P-value <0.05 (two-sided) was considered significant. EFS and OS were defined as the times from diagnosis to event (any death, relapse, secondary malignancy or failure of therapy) and from diagnosis to death from any cause or to the last follow-up, respectively. Patients without an event of interest were censored at the date of last contact. Hazard ratios for probability of relapse between subgroups were calculated using univariate Cox models. Other comparisons were performed using the χ2 test or Fisher exact test, as appropriate.
Results
Identification of kinase fusions in BCP-ALL
Twenty-nine patients with kinase fusions were identified (Table 1, Supplementary Table S3), of whom 16 were identified by mRNA-seq and 13 by mRT-PCR. All kinase fusions identified by mRNA-seq were confirmed to be in-frame with intact tyrosine kinase domain by RT-PCR and Sanger sequencing. The involved exons in each kinase fusion except CRLF2-related ones and IgH-EPOR are listed in Supplementary Table S4. It was likely that mRNA-seq was more sensitive to detect the kinase fusion (16 of the 109, 14.7% by mRNA-seq vs 13 of the 264, 4.9% by mRT-PCR), simply because only mRNA-seq can detect a novel kinase fusion. However, comparing the detection frequency of the 15 kinase fusions that were included in the mRT-PCR system, we identified 9 of the 109 (8.3%) by mRNA-seq and 13 of the 264 (4.9%) by mRT-PCR assay. Therefore, the sensitivity of two detection methods is not significantly different (9/109 vs 13/264, P=0.32).
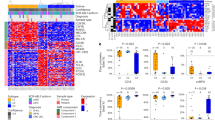
Regarding chimeric fusions, three patients had ABL1 rearrangements (SNX2, ZMIZ1 and ETV6), eight had PDGFRB rearrangements (EBF1 in six patients and ATF7IP18 in two patients), two had JAK2 rearrangements (PAX5 and OFD120), three had IgH-EPOR and one had NCOR1-LYN.20 Of the 14 patients with CRLF2 rearrangements (P2RY8 in 11 patients, IgH in 2 patients and CSF2RA21 in 1 patient), 2 harbored IgH-EPOR and ATF7IP-PDGFRB (Table 1). MLPA analysis detected IKZF1 deletions in 16 of the 22 (72.7%) patients (Table 2, Supplementary Table S3). Mutational analysis of JAK2 was performed in 12 of the 14 patients with CRLF2 rearrangement and identified 2 of the 12 (16.7%) patients with JAK2 R683-activating mutations (Table 2). The results of MLPA analysis in 29 kinase fusion-positive patients are summarized in Supplementary Table S3. Gene set enrichment analysis revealed that gene expression profile of the patients harboring kinase fusion except for P2RY8-CRLF2 was similar to that of BCR-ABL1-positive patients (Figure 2).
Clinical characteristics and outcomes of BCP-ALL patients with kinase fusions
Analysis of the clinical characteristics of the 29 patients showed male predominance (male/female: 22/7), older age at onset (median age at diagnosis: 8.8 years) and high white blood cell (WBC) count at diagnosis (median WBC count at diagnosis: 94 200/μl), which reflected the predominance of NCI-HR patients (NCI-standard risk (SR)/HR: 8/21) (Table 3). Thirteen of the 29 (44.8%) patients showed poor response to initial prednisolone (PSL) therapy. Ten patients (34.5%) who received allo-HSCT were in the first complete remission (CR), and six of them (60%) maintained first CR. Four patients (13.8%) did not respond to the initial treatment, and two of them were alive in CR by allo-HSCT. The remaining two patients died of treatment-related complications, including transplant-related mortality. Eleven patients (37.9%) experienced relapse after first CR, of whom six (54.5%) died; two of the six patients had received allo-HSCT after relapse. Only one patient who harbored SNX2-ABL1 was treated with tyrosine kinase inhibitors, such as imatinib and dasatinib; the patient did not respond well to tyrosine kinase inhibitors in combination with chemotherapy at the first and second relapse.16 The clinical course of 29 patients is summarized in Supplementary Figure S1.
In the survival analysis, the 5-year EFS and OS rates were 48.6±9.7% and 73.5±8.6%, respectively, with a median follow-up period of 6.7 years (Figure 3a). To determine prognostic significance of kinase-activating fusions, we compared 5-year EFS between kinase fusion-positive (n=16) and -negative (n=93) in mRNA-seq cohort. The 5-year EFS rate was 41.0±13.0% in the kinase-activating fusion-positive group and 67.0±7.7% in the fusion-negative group, respectively. The prognosis of kinase-activating fusion-positive patients was significantly inferior to that of kinase fusion-negative patients (log-rank P=0.035). When mRNA-seq cohort was split into two groups based on the clinical trial, such as TCCSG (n=92) and JACLS cohorts (n=17), the 5-year EFS rate was 38.1±19.9% in activating kinase fusion-positive patients and 68.9±9.3% in activating kinase fusion-negative patients in the TCCSG cohort. The difference was statistically significant (log-rank P=0.039). On the other hand, the 5-year EFS was 50.0±20.4% in activating kinase fusion-positive B-others patients and 54.6±15.0% in negative B-others patients in the JACLS cohort. The difference was not statistically significant (log-rank P=0.89). Because all of the 17 patients in the JACLS cohort have IKZF1 deletion, the prognosis of these patients was poor irrespective of the presence of kinase-activating fusions. According to the NCI risk classification, the 5-year EFS rate was 57.1±18.7% in the SR group and 44.4±11.2% in the HR group. The 5-year OS rate was 85.7±13.2% in the SR group and 65.8±10.5% in the HR group (Figures 3b and c). Although univariate analysis was performed to determine the factors related to inferior EFS or OS in 29 patients, none of the covariates such as age at diagnosis, WBC count at diagnosis, NCI risk, initial PSL response, IKZF1 deletion or allo-HSCT in first CR were statistically significant (Table 4).
According to the type of chimeric fusions, among patients with ABL-class rearrangements, such as ABL1, PDGFRB and LYN, which could be targets of imatinib or dasatinib, the 5-year EFS and OS rates were 63.6±14.5% and 90.0±9.5%, respectively (Supplementary Figures S2A and B). Among patients with JAK-class rearrangements, such as JAK2, CRLF2 and EPOR, which could be targets of JAK2 inhibitors, the 5-year EFS and OS rates were 37.5±12.1% and 62.5±12.1%, respectively (Supplementary Figures S2A and B). There were no significant differences in EFS and OS between the two genetic subgroups (log-rank P=0.42 and 0.18, respectively). No significantly different factors, such as age at onset, initial WBC count, NCI risk, IKZF1 deletion, initial PSL response and the number of patients receiving allo-HSCT in first CR, were observed between the two genetic subgroups (Table 5).
Discussion
Roberts et al.7 identified kinase fusions in 172 of the 264 patients with Ph-like ALL using mRNA-seq. Although 106 (61.6%) of the 172 patients harbored CRLF2-related fusions (45 P2RY8-CRLF2 and 61 IgH-CRLF2), the chimeric fusions identified in the remaining 66 patients varied. In detail, 36 (20.9%) patients harbored ABL-class rearrangements, including ABL1, ABL2, CSF1R and PDGFRB; 28 (16.3%) patients harbored EPOR or JAK2 rearrangement; and the remaining 2 (1.2%) patients harbored rare kinase fusions, such as ETV6-NTRK3 and ZFAND3-DGKH. IKZF1 deletion was present in 45 (68.2%) of the 66 patients with kinase fusions, excluding CRLF2-related fusions. In addition, 68 (55%) of the 106 patients with CRLF2 rearrangement harbored JAK2-activating mutation.
In the present study, we analyzed 373 patients with BCP-ALL without hyperdiploid karyotype and recurrent fusions, such as ETV6-RUNX1, E2A-PBX1, MLL-related fusions and BCR-ABL, because we did not have enough data for gene expression profiling, which is mandatory for the diagnosis of Ph-like ALL. We identified 29 kinase fusion-positive patients: 14 patients with CRLF2 rearrangements, 12 patients with ABL-class rearrangements, and 5 patients with EPOR or JAK2 rearrangement. Our retrospective analysis confirmed three types of kinase rearrangements, including CRLF2, ABL1-class and JAK2/EPOR, which were dominant, although we may have missed several patients with kinase fusions owing to lack of data in 264 patients who were not analyzed by mRNA-seq. A high prevalence (16 of the 22, 72.7%) of IKZF1 deletion in these patients was also confirmed in our study, suggesting that kinase fusion-positive BCP-ALL patients are biologically similar to Ph+ ALL patients.22 However, JAK2 mutation was present in only 2 (16.7%) of the 12 patients with CRLF2 rearrangements, which was a lower rate than that (55%) reported in previous studies.5 The reason for this discrepancy is unclear. Because other genetic alterations related to the JAK-STAT pathway, such as activating mutations in the IL7R, FLT3, JAK1 and JAK3 genes and the deletion of SH2B3, were not investigated in our study, these genetic alterations may have replaced JAK2-activating mutations in our cohort.6, 7 Additional comprehensive genetic studies are warranted to identify the therapeutic targets in our cohort.
We confirmed that the clinical characteristics of patients with kinase fusions are those commonly associated with Ph+ and Ph-like ALL, such as older age at diagnosis, male predominance and high WBC count at diagnosis.6, 7, 23, 24 In terms of treatment response, we showed that the prevalence of PSL poor responders was high in this subgroup, compared with the findings of the BFM2000 study (44.8% vs 6.3%, P<0.01).25 Compared with the kinase-activating fusion-negative patients in this study, the rate of induction failure was also high in this subgroup (12.9% vs 2.1%, P=0.004). However, the PSL poor responders and induction failure rates in our study did not differ significantly from those reported in the Ph+ ALL cohort (44.8% vs 21.4%, P=0.07 and 13.8% vs 14%, P=0.86, respectively).24 These findings suggested clinical similarities between Ph+ ALL and BCP-ALL with kinase fusions other than BCR-ABL.
The prognostic impact of Ph-like signature is controversial.7, 10, 23, 26 In addition, prognostic significance of kinase-activating fusions should be determined. In the present study, the 5-year EFS rates of kinase fusion-positive patients was significantly inferior to those of kinase fusion-negative patients in the mRNA-seq cohort, which included non-biased B-others (log-rank P=0.035). The limitation of this study was that mRNA-seq was performed only in 109 of the 373 (29.2%) patients. Complete genetic study of larger cohort is needed to clarify the prognostic significance of kinase-activating fusions in pediatric BCP-ALL.
Although information about risk factors associated with Ph-like ALL is limited, Roberts et al.7, 10 suggested that age at onset and minimal residual disease status are related to poor prognosis. In the present study, the small sample size prevented identification of risk factors related to inferior EFS and OS in our cohort, such as age at diagnosis, NCI risk and the presence of IKZF1 deletion. Furthermore, we focused on analyzing differences in the biological or clinical characteristics between genetic subgroups, such as those with ABL-class rearrangements and JAK-class rearrangements. Although EFS and OS tend to be low among patients with JAK-class rearrangements, there were no statistically significant differences between groups (data not shown). Furthermore, we could not find any difference in clinical and biological features between the two genetic subgroups (Table 5). However, our study was limited by its small sample size, its retrospective nature, the heterogeneity of treatment protocols and no data of minimal residual disease status. Thus further studies using larger cohorts are required to determine the prognostic factors in this unique subgroup. International collaborating studies are warranted in the near future.
Finally, the rapid identification of tyrosine kinase inhibitor-eligible patients is mandatory. The establishment of systematic screening systems is currently in progress in the pediatric study groups. For example, the Children’s Oncology Group used a low-density gene expression array to identify Ph-like ALL.7 We are currently developing fluorescence in situ hybridization protocols for the detection of ABL1, PDGFRB and JAK2 rearrangements, which are targetable and relatively frequent genetic abnormalities. In the future, efficient screening systems for the identification of rare but treatable kinase fusions should be established to enable the design of tailored therapy protocols for patients with high-risk BCP-ALL.
References
Inaba H, Greaves M, Mullighan CG . Acute lymphoblastic leukemia. Lancet 2013; 381: 1943–1955.
Schultz KR, Carroll A, Heerema NA, Bowman WP, Aledo A, Slayton WB et al. Long-term follow-up of imatinib in pediatric Philadelphia chromosome-positive acute lymphoblastic leukemia: Children’s Oncology Group Study AALL0031. Leukemia 2014; 28: 1467–1471.
Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009; 360: 470–480.
Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 2013; 45: 242–252.
Chen IM, Harvey RC, Mullighan CG, Gastier-Foster J, Wharton W, Kang H et al. Outcome modeling with CRLF2, IKZF1, JAK, and minimal residual disease in pediatric acute lymphoblastic leukemia: a Children's Oncology Group study. Blood 2012; 119: 3512–3522.
Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 2012; 22: 153–166.
Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang D, Pei K et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 2014; 371: 1005–1015.
Weston BW, Hayden MA, Roberts KG, Bowyer S, Hsu J, Fedoriw G et al. Tyrosine kinase inhibitor therapy induces remission in a patients with refractory EBF1-PDGFRB-positive acute lymphoblastic leukemia. J Clin Oncol 2013; 31: e413–e416.
Lengline E, Beldjord K, Dombret H, Soulier J, Boissel N, Clappier E . Successful tyrosine kinase inhibitor therapy in a refractory B-cell precursor acute lymphoblastic leukemia with EBF1-PDGFRB fusion. Haematologica 2013; 98: e146–e148.
Roberts KG, Pei D, Campana D, Payne-Turner D, Li Y, Chen C et al. Outcomes of children with BCR-ABL-like acute lymphoblastic leukemia treated with risk-directed therapy based on the levels of minimal residual disease. J Clin Oncol 2014; 32: 3012–3020.
Hasegawa D, Hara J, Suenobu S . Successful abolition of prophylactic cranial irradiation in children with non-T acute lymphoblastic leukemia (ALL) in the Japan Association of Childhood Leukemia Study (JACLS) ALL-02 trial. Blood (ASH Annual Meeting Abstracts) 2011; 118: 653.
Suzuki N, Yumura-Yagi K, Yoshida M, Hara J, Nishimura S, Kudo T et al. Outcome of childhood acute lymphoblastic leukemia with induction failure treated by the Japan Association of Childhood Leukemia study (JACLS) ALL F-protocol. Pediatr Blood Cancer 2010; 54: 71–78.
Kato M, Manabe A, Koh K, Inukai T, Kiyokawa N, Fukushima T et al. Treatment outcome of adolescent acute lymphoblastic leukemia treated on Tokyo Children’s Cancer Study Group (TCCSG) clinical trials. Int J Hematol 2014; 100: 180–187.
Hyakuna N, Shimomura Y, Watanabe A, Taga T, Kikuta A, Matsushita T et al. Assessment of corticosteroid-induced osteonecrosis in children undergoing chemotherapy for acute lymphoblastic leukemia: a report from the Japanese Childhood Cancer and Leukemia Study Group. J Pediatr Hematol Oncol 2014; 36: 22–29.
Okamoto Y, Koga Y, Inagaki J, Ozono S, Ueda K, Shimoura M et al. Effective VCR/DEX pulse maintenance therapy in the KYCCSG ALL-02 protocol for pediatric acute lymphoblastic leukemia. Int J Hematol 2016; 103: 202–209.
Masuzawa A, Kiyotani C, Osumi T, Shioda Y, Iijima K, Tomita O et al. Poor responses to tyrosine kinase inhibitors in a child with precursor B-cell acute lymphoblastic leukemia with SNX2-ABL1 chimeric transcript. Eur J Haematol 2014; 92: 263–267.
McPherson A, Hormozdiari F, Zayed A, Guiliany R, Ha G, Sun MG et al. R deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 2011; 7: e1001138.
Kobayashi K, Mitsui K, Ichikawa H, Nakabayashi K, Matsuoka M, Kojima Y et al. ATF7IP as a novel PDGFRB fusion partner in acute lymphoblastic leukaemia in children. Br J Haematol 2014; 165: 836–841.
Asai D, Imamura T, Suenobu S, Saito A, Hasegawa D, Deguchi T et al. IKZF1 deletion is associated with a poor outcome in pediatric B-cell precursor acute lymphoblastic leukemia in Japan. Cancer Med 2013; 2: 412–419.
Yano M, Imamura T, Asai D, Kiyokawa N, Nakabayashi K, Matsumoto K et al. Identification of novel kinase fusion transcripts in pediatric B cell precursor acute lymphoblastic leukaemia with IKZF1 deletion. Br J Haematol 2015; 171: 813–817.
Yano M, Imamura T, Asai D, Moriya-Saito A, Suenobu S, Hasegawa D et al. An overall characterization of pediatric acute lymphoblastic leukemia with CRLF2 overexpression. Genes Chromosomes Cancer 2014; 53: 815–823.
Mullighan CG, Miller CB, Radtke I, Phillips LA, Dalton J, Ma J et al. BCR-ABL1 lymphoblastic leukemia is characterized by the deletion of Ikaros. Nature 2008; 453: 110–114.
Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JGCAM, Peters STCJM et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol 2009; 10: 125–134.
Manabe A, Kawasaki H, Shimada H, Kato I, Kodama Y, Sato A et al. Imatinib use immediately before stem cell transplantation in children with Philadelphia chromosome-positive acute lymphoblastic leukemia: results from Japanese Pediatric Leukemia/Lymphoma Study Group (JPLSG) Study Ph+ALL04. Cancer Med 2015; 4: 682–689.
Conter V, Bartram CR, Valsecchi MG, Schrauder A, Panzer-Grumayer R, Moricke A et al. Molecular response to treatment redefines all prognostic factors in children and adolescents with B-cell precursor acute lymphoblastic leukemia: results in 3184 patients of the AIEOP-BFM ALL 2000 study. Blood 2010; 115: 3206–3214.
van der Veer A, Waanders E, Pieters R, Willemse ME, van Reijmersdal SV, Russell LJ et al. Independent prognostic value of BCR-ABL1 like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood 2013; 122: 2622–2629.
Acknowledgements
This work was supported by grants for Clinical Cancer Research and Research on Measures for Intractable Diseases from the Japanese Ministry of Health, Labor and Welfare, by grants-in-aid for scientific research from the Japanese Ministry of Education, Culture, Sports, Science and Technology and by the Practical Research for Innovative Cancer Control from Japan Agency for Medical Research and development, AMED (15ck0106066h0002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Blood Cancer Journal website
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Imamura, T., Kiyokawa, N., Kato, M. et al. Characterization of pediatric Philadelphia-negative B-cell precursor acute lymphoblastic leukemia with kinase fusions in Japan. Blood Cancer Journal 6, e419 (2016). https://doi.org/10.1038/bcj.2016.28
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bcj.2016.28
This article is cited by
-
Philadelphia-like acute lymphoblastic leukemia: the journey from molecular background to the role of bone marrow transplant—review article
Annals of Hematology (2023)
-
A reporter system for enriching CRISPR/Cas9 knockout cells in technically challenging settings like patient models
Scientific Reports (2021)
-
Hematological characteristics, cytogenetic features, and post-induction measurable residual disease in thymic stromal lymphopoietin receptor (TSLPR) overexpressed B-cell acute lymphoblastic leukemia in an Indian cohort
Annals of Hematology (2021)
-
Philadelphia chromosome-negative B-cell acute lymphoblastic leukaemia with kinase fusions in Taiwan
Scientific Reports (2021)
-
Risk-adjusted therapy for pediatric non-T cell ALL improves outcomes for standard risk patients: results of JACLS ALL-02
Blood Cancer Journal (2020)