Abstract
Background:
In a clinical diagnostic laboratory, we evaluated the applicability of the Ion Proton sequencer for screening 409 cancer-related genes in solid tumours.
Methods:
DNA was extracted from formalin-fixed, paraffin-embedded (FFPE) tissue biopsy specimens of 55 solid tumours (20 with matched normal tissue) and four cell lines and screened for mutations in 409 genes using the Ion Proton system. The mutation profiles of these samples were known based on prior testing using the Ion Torrent Personal Genome Machine (46-gene hotspot panel), Sanger sequencing, or fluorescence in situ hybridisation (FISH). Concordance with retrospective findings and additional mutations were evaluated. Assay sensitivity and reproducibility were established. Gene copy number variations (CNVs) detected were confirmed by molecular inversion probe (MIP) array.
Results:
The average Ion Proton (409-gene panel) sequencing output per run was 8 gigabases with 128 million sequencing reads. Of the 15,992 amplicons in the 409-gene panel, 90% achieved a minimum average sequencing depth of 100X. In 59 samples, the Ion Proton detected 100 of 105 expected single-nucleotide variants (SNVs) and all expected deletions (n=8), insertions (n=5), and CNVs (n=7). Five SNVs were not detected due to failed amplification of targeted regions. In 20 tumours with paired normal tissue, Ion Proton detected 37 additional somatic mutations, several in genes of high prognostic or therapeutic significance, such as MET, ALK, TP53, APC, and PTEN. MIP array analysis confirmed all CNVs detected by Ion Proton.
Conclusions:
The Ion Proton (409-gene panel) system was found to be well suited for use in a clinical molecular diagnostic laboratory. It can simultaneously screen 409 genes for a variety of sequence variants in multiple samples using a low input of FFPE DNA with high reproducibility and sensitivity.
Similar content being viewed by others
Main
Comprehensive screening of genetic abnormalities in tumours has become an important part of the clinical workup of cancer patients. The detection of specific genetic aberrations can facilitate prognostic stratification and guide the selection of targeted therapies. Until recently, routine screening of tumours for mutations was confined to low- and medium-throughput platforms. More recently, massively parallel and deep sequencing of the cancer genome using next-generation sequencing (NGS) technologies has vastly increased the rate at which clinically significant and therapeutically targetable mutations are discovered (Ross and Cronin, 2011; Dutton-Regester and Hayward, 2012; Li et al, 2013; Martelli et al, 2013; Mwenifumbo and Marra, 2013; Workman et al, 2013). Consequently, the number of genes that warrant routine screening in a clinical molecular diagnostic laboratory has increased considerably. However, most sequencing platforms currently validated for clinical use, such as Sanger sequencing, pyrosequencing, and primer extension coupled with mass spectroscopy, are limited in their ability to assess multiple markers simultaneously using small amounts of DNA. These limitations can be potentially circumvented by the introduction of NGS platforms into the clinical laboratory that can provide high throughput and multiplexed sequencing capabilities.
In recent years, a few clinical molecular laboratories have applied and validated NGS platforms for routine diagnostic screening of solid tumours (Beadling et al, 2013; Singh et al, 2013; Spencer et al, 2013). Although a paradigm shift for mutation detection is already well under way, the number of genes being routinely screened in clinically validated NGS assays is still relatively low (20–50 genes) and generally restricted to well-established markers of clinical or therapeutic significance (e.g., EGFR, KRAS, NRAS, TP53, etc.). Additionally, these assays are focused on hotspot mutation regions and were designed to detect single-nucleotide variations (SNVs), insertions, and deletions but not copy number variations (CNVs). Furthermore, owing to the large number of genetic abnormalities being identified in virtually every tumour type, there is a pressing need to incorporate higher-throughput NGS platforms to comprehensively screen large numbers of genes. To meet this need, testing the clinical applicability of newer high-capacity NGS platforms is warranted. Towards this goal, we validated a commercially available NGS panel designed to screen all exons of 409 cancer-related genes using Ion Proton (Life Technologies, Carlsbad, CA, USA), a high-capacity sequencer that employs a semiconductor-based technology interfaced with sequencing-by-synthesis that is similar to its lower-capacity counterpart, the Ion Torrent Personal Genome Machine (IT-PGM) (Life Technologies) (Rothberg et al, 2011).
In a Clinical Laboratory Improvement Amendment certified laboratory, we tested the sensitivity, specificity, reproducibility, and applicability of using the Ion Proton 409-gene panel to routinely screen for SNVs, insertions/deletions, and CNVs using as little as 60 ng of DNA extracted from formalin-fixed, paraffin-embedded (FFPE) tumour tissue. Our results show the utility of using this approach to assess mutations and CNVs. Importantly, the Ion Proton instrument can detect abnormalities in real time with less than a week turnaround time from the receipt of sample and therefore can be used to stratify patients into prognostic groups and potentially guide therapeutic decisions.
Materials and methods
Tumour samples
We used 55 tumours (20 with paired normal tissues) and four cancer cell lines for sequencing studies. The tumours selected were diverse and included cases of melanoma, gastrointestinal stromal tumours, brain tumours, and carcinomas of the lung, breast, gynaecologic tract, and gastrointestinal tract. The age of the FFPE tissue biopsy samples ranged from 1 to 8 years. This study was approved by the Institutional Review Board (IRB protocol PA13-0512). The cell lines included H2122 (CRL-5985), H460 (HTB-177), DLD1 (CCL221), and HL60 (CCL240) purchased from ATCC (Manassas, VA, USA). The cell lines were fixed in formalin and embedded in paraffin to mimic routinely processed tissue biopsy specimens. All tumour samples and cell lines used in this study had been tested previously using either the IT-PGM (46-gene panel), Sanger sequencing, or fluorescence in situ hybridisation (FISH).
Tissue selection and DNA extraction
Hematoxylin and eosin stained tissue sections of FFPE tumour biopsy samples were reviewed and the tumour area was circled by a pathologist. Only specimens with a minimum of 20% tumour in the circled area were selected for this study. Tumours with high degree of necrosis and less than 1000 tumour cells in the circled area were not included. Unstained tissue sections of 5 μM thickness were deparaffinised and manually microdissected using the hematoxylin- and eosin-stained slide as a guide. DNA extraction and purification were performed using the PicoPure DNA extraction kit (Arcturus, Mountain View, CA, USA) and Agentcourt AMPureXP kit (Agentcourt Biosciences, Beverly, MA, USA), respectively. The Qubit DNA HS assay kit (Life Technologies) was used to quantify purified DNA.
Library preparation for the Ion Proton 409-gene assay
Library preparation for the 409-gene panel was performed using the Ion Torrent Ampliseq Comprehensive Cancer Panel (Life Technologies) following the manufacturer’s instructions. The panel was designed to facilitate amplification-based capture and sequencing of coding regions of 409 cancer-related genes. It includes four primer pools with approximately 4000 primer pairs in each pool and requires a total of 60 ng of DNA as a template for each sample (15 ng per pool). Sample barcoding was accomplished using the Ion Xpress Barcode Adapter 1–96 kit (Life Technologies). The library prepared in each pool was quantified using the 2200 TapeStation System (Agilent Technologies, Santa Clara, CA, USA).
Automated emulsion PCR and sequencing of 409-gene panel using Ion Proton
To clonally amplify the 409-gene panel DNA library on Ion Spheres (Life Tecnologies), emulsion polymerase chain reaction (PCR) was performed using the Ion PI Template OT2 kit v2 and Ion One Touch 2 system (Life Technologies) as per the manufacturer’s instructions and as described in detail in the Supplementary Methods.
Sequencing data analysis using Ion Proton
Alignment of sequencing reads with the reference genome and base calling was performed using Torrent Suite software v3.6.2 (Life Technologies). Human genome build 19 was used as the reference for alignment. Identification of sequence variants was facilitated via IT Variant Caller Plugin software v3.6.59049 (Life Technologies) and coverage of each amplicon was obtained by the Coverage Analysis Plugin software v3.6.58977 (Life Technologies). The Integrative Genomics Viewer (IGV) (Thorvaldsdottir et al, 2012) was used to visualise the read alignment and the presence of variants against the reference genome as well as to confirm variant calls by checking for strand biases and sequencing errors. A custom in-house developed software (OncoSeek) described earlier (Singh et al, 2013) was used to interface the data generated by the Ion Torrent Variant Caller (Life Technologies) with the IGV, filter repeat errors due to nucleotide homopolymer regions, compare replicate samples, and annotate the sequencing information to generate a clinical report. Torrent Suite v4.0.2 (Variant Caller plugin v4.0-r76860 and Coverage Analysis plugin v4.0-r77897) (Life Technologies) was used to reanalyse the sequencing data for a subset of samples.
Sensitivity analysis of the Ion Proton 409-gene panel assay
Variant detection sensitivity was analysed by sequencing sequentially diluted DLD1 cell line (FFPE DNA) into DNA from H460 cell line (FFPE DNA) in ratios of 1:3, 1:9, 1:19, and 1:39, resulting in 25, 10, 5, and 2.5% dilutions, respectively. Confidence intervals were estimated using the efficient-score method corrected for continuity (Newcombe, 1998).
Intra-run and inter-run reproducibility of the Ion Proton 409-gene panel assay
Five distinct barcoded samples of 25% DLD1 DNA (1:3 dilution of DLD1 in H460) were multiplexed and sequenced in the same run to assess the intra-run reproducibility of sequencing on the Ion Proton. To determine the inter-run reproducibility, 25% DLD1 DNA was sequenced in 10 independent runs. In both analyses, the ability of the platform to detect mutations at the expected variant frequencies was assessed.
Library preparation and sequencing using IT-PGM (46-gene panel)
The Ion Ampliseq Cancer Panel (Life Technologies) covering mutation hotspot regions of 46 genes was used for library preparation and sequencing using the IT-PGM sequencer (Life Technologies) as described previously (Singh et al, 2013). Briefly, starting from 10 ng FFPE DNA, library preparation was performed using an Ion Ampliseq 2.0 kit and IT Ampliseq Cancer Panel primers (Life Technologies). From the barcoded library, manual emulsion PCR was performed, followed by sequencing eight multiplexed samples using Ion 318 chips and the IT-PGM Torrent Suite v2.0 (Life Technologies) was used for analysis.
Comparison of Ion Proton and the MiSeq sequencers
For comparison, FFPE DNA from three cell lines (DLD1, H460, and H2122) sequenced using Ion Proton 409-gene panel were also subjected to sequencing using the modified hotspot 54-gene TruSeq Amplicon Cancer Panel and MiSeq sequencer (Illumina, San Diego, CA, USA). Starting from 100 ng of DNA, MiSeq 54-gene panel library preparation and sequencing were performed according to the manufacturer’s protocol as described previously (Luthra et al, 2014). Mutations detected by the MiSeq (54-gene panel) were compared to the mutation detected by Ion Proton 409-gene panel.
CNV detection using the Ion Proton 409-gene panel assay
The presence of CNVs was assessed by comparing the average sequencing depth achieved for amplicons covering a gene in a sample to the average sequencing depth achieved historically for those amplicons in all the samples (n=300) in our database, normalised against the sequencing efficiency of the sample (total number of AQ20 reads for the sample) following an algorithm described in Supplementary Methods.
Sensitivity for CNV detection using the Ion Proton 409-gene panel assay
CNV detection sensitivity was assessed by serially diluting FFPE H460 cell line DNA into DNA extracted from FFPE normal brain tissue to obtain H460 dilutions of 100, 50, 25, 12.5, and 6.25%. Samples were tagged with different barcodes and sequenced on the same chip. Two separate estimations of sensitivity were performed and compared.
CNV analysis by molecular inversion probe array
Genome-wide CNV analysis via molecular inversion probe (MIP) array was performed using the OncoScan FFPE assay kit (Affymetrix, Santa Clara, CA, USA) following the manufacturer’s recommended protocol as described in Supplementary Methods.
Fluorescence in situ hybridisation (FISH) analysis
The MYC gene copy number was assessed by FISH using a break-apart probe (Abbott Molecular, Abbott Park, IL, USA) mixed with a single-colour (aqua) MYB probe (chromosome 6q23) as an internal control. EGFR FISH was performed using a dual-colour FISH probe set (Abbott Molecular) containing a centromeric probe for chromosome 7 as an internal control. 200 cells were counted for each probe.
Results
Sequencing metrics
An average sequencing output of 8 gigabases (Gb) was obtained for 26 sequencing runs. This output represented an average of 128.5 million sequencing reads per run, of which an average of 32% of reads (40.9 million) were polyclonal and 8% of reads (7.6 million) were of low quality and filtered. Therefore, on average in every sequencing run 80 million reads were of high quality (⩾ AQ20 or one error in 100 base pairs (bp)) and provided useful sequence information. The overall performance of the amplicons as determined by their average sequencing depth is provided in Supplementary Table 1.
Sensitivity studies
The sensitivity of the Ion Proton (409-gene panel) platform for detecting SNVs was estimated by using DLD1 cell line FFPE DNA sequentially diluted into H460 cell line FFPE DNA. DLD1 harbours heterozygous mutations in seven different genes: IDH1, PIK3CA, KRAS, KIT, MYD88, TP53, and SMO. The ability of the platform to detect these mutations in 50%, 25%, 10%, and 5% dilutions of DLD1 DNA was assessed in two different sequencing runs performed on different Ion Proton sequencers. In both runs, all expected mutations were called in 100 and 50% of diluted DLD1 DNA. In the 25% dilution, an SMO mutation was missed in one run; otherwise, all the expected mutations were detected in both runs. Similarly, in the 10% dilution of DLD1, the SMO mutation was missed in one run, whereas the remaining mutations were consistently detected in both runs. In the 5% dilution of DLD1, multiple mutations were not detected in both runs. Of interest, two mutations (TP53 and SMO) that were missed in the 25 and 10% dilutions of DLD1 were covered by amplicons that were sequenced at a relatively low sequencing depth (<200 × ). These mutations were evident in the sequencing reads; however, owing to low sequencing depth, they were not called by the variant caller software (TS v3.6.2). A detailed summary of the dilution studies performed with the expected and detected levels at each dilution and the associated linearity, sensitivity, and confidence interval are provided in Supplementary Table 2.
Inter-run and intra-run reproducibility
Inter-run reproducibility was determined by sequencing a 25% dilution of DLD1 cell line FFPE DNA into H460 cell line FFPE DNA across five different sequencing runs. The results showed that each expected mutation was detected in every run with little variation in the allelic fractions (Supplementary Figure 1, upper panel). Similarly, intrarun reproducibility was established by sequencing five aliquots of 25% DLD1 cell line FFPE DNA, each tagged with distinct barcodes on the same sequencing run. The results showed consistent detection of mutations at comparable allelic fraction levels, indicating high intrarun sequencing reproducibility (Supplementary Figure 1, lower panel).
Concordance of Ion Proton (409-gene panel) sequencing with IT-PGM (46-gene panel) and Sanger sequencing
In four cell lines and the set of 28 tumours without paired normal tissue, a total of 81 sequence variants were expected. Seventy-seven sequence variants (69 SNVs, six deletions, and two insertions) were expected based on sequencing using the IT-PGM (46-gene panel) and four sequence variants based on Sanger sequencing (three insertions and one deletion). The Ion Proton 409-gene panel sequencing detected 64 of 69 expected SNVs. Five SNVs were not detected owing to target amplification failure, including three samples in which there was failure of the same amplicon (samples 2, 12, and 33, Supplementary Table 3). Of note, in the DLD1 cell line, the Ion Proton (409-gene panel) detected 4 SNVs not detected by the IT-PGM attributable to the fact that the 46-gene hotspot panel lacked sequencing coverage in these areas.
In a melanoma sample (Supplementary Table 3, sample 4), the IT-PGM (46-gene panel) detected a BRAF p.V600K (c.GTG>AAG) mutation at a 69% allelic fraction (Figure 1). This mutation was also detected by the Ion Proton (409-gene panel) at an 81% allelic fraction (Figure 1A). In a second example (colon adenocarcinoma, Supplementary Table 3, sample 15), a KRAS p.G12S mutation was detected by the IT-PGM and was also detected by the Ion Proton at a comparable allelic fraction (Figure 1B). In another instance (colon adenocarcinoma, Supplementary Table 3, sample 18), a PDGFRA mutation (p.P553L, c.CCG>CTG) was detected by the IT-PGM at a 29% allelic fraction and was also detected by the Ion Proton at a 20% allelic fraction (Figure 1C).
Concordance of SNV detection between Ion Proton (409-gene panel) and IT-PGM (46-gene panel). Examples showing the concordance of mutation detection between Ion Proton (409-gene panel) and IT-PGM (46-gene panel) are presented. (A) IT-PGM detected a dual mutation in BRAF (p.V600K) in a melanoma sample that was also detected by Ion Proton. The nucleotide change was GTG>AAG. The BRAF gene has a reverse orientation on chromosome 7 (direction indicated by the arrow). However, the IGV exhibits aligned reads only in the ‘forward’ orientation which makes the substituted nucleotide appear as TT instead of AA (CAC>TTC or GAG>AAG). (B) IT-PGM detected a KRAS (p.G12S, c.GGT>AGT) mutation in a colon adenocarcinoma sample that was also detected by Ion Proton at a comparable allelic fraction. As KRAS is also located in the reverse orientation on chromosome 12 (indicated by the arrow) the substituted base appears in IGV as ‘T’ instead of ‘A’ (CCA>TCA or GGT>AGT) (C) A PDGFRA (p.P553L, c.CCG>CTG) mutation was detected by IT-PGM (46-gene panel) in a colon adenocarcinoma sample, which was also detected by Ion Proton (409-gene panel).
To assess the ability of the Ion Proton (409-gene panel) to detect deletions, we tested a set of seven tumours including five adenocarcinomas (two lung, two colon, and one gynaecological tract) and two gastrointestinal stromal tumours with known deletions as detected by the IT-PGM (46-gene panel) (n=5) or by Sanger sequencing (n=2). The Ion Proton (409-gene panel) detected all seven deletions expected in these samples (Supplementary Table 3). Examples of two deletions (a 15-bp deletion in exon 19 of EGFR and a 5-bp deletion in exon 16 of APC) detected by both the IT-PGM and Ion Proton are shown in Figure 2.
Detection of deletions by Ion Proton (409-gene panel). Samples with known deletions as detected by IT-PGM (46 gene panel) were also sequenced on Ion Proton (409-gene panel) and compared (A) A 15-bp deletion detected in exon 19 of EGFR in a lung adenocarcinoma sample by IT-PGM (46-gene panel) was also detected by Ion Proton (409-gene panel). (B) A 5-bp deletion in exon 16 of APC detected in a colon adenocarcinoma sample by IT-PGM (46-gene panel) was also detected by Ion Proton (409-gene panel).
Similarly, the ability of the Ion Proton (409-gene panel) to detect insertions was tested by sequencing five samples (three lung adenocarcinomas, one brain tumour, and one gastrointestinal stromal tumour) with known insertions as determined previously by the IT-PGM (46-gene panel) (n=2) or Sanger sequencing (n=3). The Ion Proton (409-gene panel) detected three of the five insertions but missed a 6-bp insertion in exon 20 of EGFR (sample 9) and a 6-bp insertion in exon 11 of KIT (samples 9 and 24, Supplementary Table 3). These insertions were evident in the sequencing reads when visualised by the IGV but were not called by the variant caller in the analysis software (v3.6.2) (Life Technologies). However, upon reanalysis of the results using a more recent version of the software (v4.0.2) (Life Technologies), both insertions were identified (Supplementary Table 4, samples 1 and 2). Representative samples with insertions detected using the IT-PGM and Ion Proton are shown in Figure 3.
Detection of insertions by Ion Proton (409-gene panel). A set of samples with known insertions as detected by Sanger sequencing was sequenced on IT-PGM (46 gene panel) and Ion Proton (409-gene panel) for comparison. In IGV, the presence of insertions in the reads is marked by the presence of the purple marker shown by the arrows. (A) A 6-bp insertion in exon 20 of EGFR was successfully detected and called by both IT-PGM (46-gene panel) and Ion Proton (409-gene panel). (B) A 6-bp insertion in KIT, exon 9 was detected and called by Ion-Proton (409-gene panel) but not by IT-PGM (46-gene panel). The insertion was evident in the IT-PGM sequencing reads but was not called by the variant caller in Torrent Suite 2.0.
Mutation detection in tumours with paired normal samples
A total of 20 tumour samples with paired normal tissue specimens were sequenced using the Ion Proton (409-gene panel). The mutational profile of these tumours was known through earlier analysis using the IT-PGM (46-gene panel) that detected 37 sequence variants (36 SNVs and one deletion). The Ion Proton detected all the SNVs except one (NRAS, p.Q61H), which was not detected owing to amplicon failure (Supplementary Table 5, sample 10). Additionally, a 2-bp deletion in TP53 (Supplementary Table 5, sample 14) was not called by the Ion Proton. This deletion was visible in the sequencing reads but was not called by the analysis software (v3.6.2). This deletion was successfully called in a subsequent analysis of the sequencing results for this sample via a later release of the software (v4.0.2) (Supplementary Table 4, sample 3). In this sample set, eight SNVs were confirmed to be germline polymorphisms present in both paired normal and tumour samples. The remaining 27 sequence variants were somatic, as they were detected in only the tumour samples (Supplementary Table 5). Representative samples showing the presence of somatic mutations in the sequencing reads of tumours, but not in paired normal tissues, are shown in Figure 4.
Detection of mutation by Ion Proton (409-gene panel) in paired normal-tumour samples. As subset of tumour samples with known mutations as detected by IT-PGM (46 gene panel) were also sequenced on Ion Proton (409-gene panel) along with their paired normal. Two representative examples with mutations detected by IT-PGM (46-gene panel) and Ion Proton (409-gene panel) in the tumours and the lack of the same in paired normal samples are shown (A) A PIK3Ca (p.E545K, c. GAG>AAG) mutation in a gastrointestinal tumour, (B) A TP53 (p.F113C, c.TTC>TGC) mutation detected in a breast carcinoma at a comparable allelic fraction.
Furthermore, to establish the concordance between software versions, we reanalysed Ion Proton (409-gene panel) sequencing results from v3.6.2 with v4.0.2 for a subset of three tumour samples and three cell lines. A complete concordance was observed; all expected sequence variants that were detected using v3.6.2 were detected using v4.0.2 (17 SNVs, two insertions, and one deletion) (Supplementary Table 4, samples 4–9).
Additional somatic mutations detected by the Ion Proton (409-gene panel) in comparison with the IT-PGM (46-gene panel)
In the 20 tumour samples with paired normal tissue specimens, additional somatic mutations were detected using the 409-gene panel on the Ion Proton compared with the IT-PGM (46-gene panel) (Supplementary Table 6). These additional mutations were identified by comparing the variants in the tumours with the lack of the same in the paired normal tissue samples and following the filtering approach explained in Supplementary Figure 2. In 16 of 20 tumour samples, 37 additional somatic mutations across 32 genes were detected (Supplementary Table 6). Seven of these mutations were detected in genes of high prognostic or therapeutic significance, including PTEN, TP53, APC, MET, and ALK, and six additional mutations were discovered in six potentially actionable genes, ERBB4, PARP1, CSF1R, SMARCA4, FBXW7, and NTRK3. Of 37 mutations detected using the Ion Proton, 10 were recorded in the COSMIC database (Forbes et al, 2008), indicating that they had been reported in the literature as somatic mutations (Supplementary Table 6). The mutations identified by the Ion Proton (409-gene panel) in TP53, APC, ERBB4, ALK, PTEN, and MET were missed by the IT-PGM (46-gene hotspot panel) owing to lack of sequencing coverage in these areas.
Concordance between Ion Proton (409-gene panel) and MiSeq (54-gene panel) sequencing results
In DNA from three cell lines (H2122, H460, and DLD1), the Ion Proton (409-gene panel) detected 17 SNVs. The MiSeq (54-gene hotspot panel) identified 15 of these 17 SNVs; the remaining two SNVs were missed owing to lack of sequencing coverage in the 54-gene panel (Supplementary Table 7). These results indicate cross-platform concordance of the Ion Proton sequencing and mutation detection with Illumina sequencing technology used in the MiSeq sequencer.
Detection of CNVs using the Ion Proton (409-gene panel)
Unlike hotspot mutation gene panels that are limited in their coverage to small frequently mutated gene regions, sequencing the entire coding sequence of a gene provides better sequencing coverage for the identification of CNVs in NGS-based assays. As all exons of the 409 genes are captured and sequenced using the Ion Proton, this method facilitates better detection of gain or loss of gene copy number. Furthermore, in a PCR-based target capture approach such as the strategy employed in this study, the number of gene copies in the genomic template influences amplification levels during target capture, which in turn is reflected in the sequencing depth. Thus, any gain or loss of copy number can be deciphered as a distinct increase or decrease in the sequencing depth achieved by the amplicons covering the gene.
CNVs were detected in three of four cell lines sequenced in our sample cohort and confirmed by the MIP array-based CNV detection assay or FISH analysis (Supplementary Table 8). For example, in the HL60 cell line, MYC amplification was detected by the Ion Proton (409-gene panel) (Figure 5A, left panel) and was confirmed by the MIP array (Figure 5A, middle and right panels). In the H460 cell line, amplification of MYC and loss of EGFR were detected by the Ion Proton (409-gene panel) (Figure 5B, top panel) and confirmed by the MIP array (Figure 5B, middle panel). FISH analysis of H460 showed monosomy of chromosome 7 and one copy of EGFR. FISH also detected 13% of the cells with monosomy of chromosome 8 with 3–6 copies of MYC, 26.5% of the cells with disomy of chromosome 8 and 3–6 copies of MYC, and 52% of the cells with apparent disomy of chromosome 8 and 10–20 copies of MYC (Figure 5B, lower panels), thereby confirming the overall MYC amplification (average, 5.6 copies) detected by the Ion Proton (409-gene panel). In the H2122 cell line, the Ion Proton (409-gene panel) detected loss of LRP1B and gain of MYC (Figure 5C, top panel) that were confirmed by the MIP array (Figure 5C, middle panels). MYC amplification detected by the Ion Proton (409-gene panel) in H2122 was also confirmed by FISH analysis, which showed 24% of cells with monosomy of chromosome 8 and 5–10 copies of MYC and 55% of cells with disomy of chromosome 8 with 10–25 copies of MYC (Figure 5C, lower panel) (average, 6.7 copies).
CNVs detected in cell lines. Ion Proton (409-gene panel) detected several CNV alterations in the three cell lines. (A) High level of sequencing depth observed for MYC in the HL60 cell line showed copy number gain also confirmed by the MIP array. (B) Gain of MYC and loss of EGFR was detected in the H460 cell line and confirmed by MIP array and FISH. (C) Amplification of MYC and loss of LRP1B were detected in the H2122 cell line and were confirmed by the MIP array. Blue and red lines adjacent to the chromosomes indicate areas of amplification and loss, respectively.
To further test the ability of the Ion Proton (409-gene panel) to detect CNVs, we sequenced four breast carcinoma samples with known ERBB2 amplification as evidenced by FISH studies. The Ion Proton (409-gene panel) successfully detected ERBB2 amplification in each sample (ERBB2 FISH positive, Supplementary Table 8, samples 1–4) and detected ERBB2 amplification in an additional tumour sample in the study group also confirmed by the MIP array (Supplementary Table 8, sample 5). In these five samples, the Ion Proton (409-gene panel) detected concurrent amplification of PGAP3 and CDK12 in addition to ERBB2. MIP array analysis also showed amplification of the locus harbouring ERBB2, PGAP3, and CDK12 (Supplementary Table 8, Figure 6A). Furthermore, in a gastrointestinal stromal tumour sample, the Ion Proton (409-gene panel) detected amplification of four genes PDGFRB, CSF1R, JAK2, and MYC (Supplementary Table 8, sample 6, Figure 6B, left panel). These amplifications were confirmed by MIP array analysis, which showed amplification of chromosome 5 locus for both PDGFRB and CSF1R along with amplification of MYC (chromosome 8) and JAK2 (chromosome 9) (Figures 6B, 3 panels on the right). In another example, amplification of EGFR was detected in a brain tumour sample by the Ion Proton and confirmed by the MIP array (Supplementary Table 8, Figure 6C). Furthermore, the copy number estimations by the Ion Proton (409-gene panel) and the MIP array were comparable (Supplementary Table 8). A summary of the concordance of Ion Proton (409-gene panel) sequencing results with results obtained using orthogonal platforms is included in Table 1.
CNV detection in patient samples. (A) ERBB2 amplifications in four breast adenocarcinoma samples positive for by FISH analysis (Samples 1–4, Supplementary Table 7) were sequenced using Ion Proton (409-gene panel), which showed the amplification of the ERBB2 along with PGAP3 and CDK12. (B) Amplifications of PDGFRB, CSF1R, JAK2, and MYC detected in a gastrointestinal stromal tumour sample were confirmed by the MIP-array. (C) EGFR copy number gain identified in a brain tumour sample was confirmed by MIP-array, which showed overall amplification of chromosome 7 including the loci for EGFR. Blue and red lines adjacent to the chromosomes indicate areas of amplification and loss, respectively.
CNV detection sensitivity
To determine the sensitivity of CNV detection, we performed a sequential dilution study using DNA from the H460 cell line (with MYC amplification and EGFR loss) diluted into DNA from a normal tissue sample to obtain H460 DNA dilutions of 50, 25, 12.5, and 6.25%. MYC amplification in H460 was consistently detected in two independent estimations in the 50, 25, and 12.5% dilutions but was undetectable in the 6.25% dilution. EGFR loss was evident in all dilutions, indicating a high sensitivity of detection for both amplifications and deletions (Supplementary Figure 3).
Discussion
Accumulation of somatic mutations and CNVs are major driving factors of oncogenesis and are potential targets for therapy. Consequently, screening tumours for genomic aberrations can be helpful for prognostic stratification and for guiding the selection of therapy. In recent years, extensive application of NGS has impacted all aspects of cancer detection and treatment, including hereditary cancer syndromes, classification of cancer subtypes, understanding of tumour heterogeneity, and identification of the genetic aberrations in problematic cases and non-responders, (Hodis et al, 2012; Liang and Kim, 2013; Roberts and Klein, 2013; Tuna and Amos, 2013; Bielinski et al, 2014; Ross et al, 2014). Novel CNVs are also being identified and can be exploited for targeted therapies (Walter et al, 2009; Bowcock, 2013). It is therefore imperative that the mutational screening of tumour DNA be extensive to assess the entire coding sequences (exons) of multiple genes to capture both sequence variants and CNVs. In this regard, NGS-based diagnostics, which are capable of detecting the full range of genetic variants, offer the potential to greatly streamline testing by using a single analysis platform. Although hotspot NGS panels that interrogate specific areas of high mutational susceptibility have been developed and are in use in some clinical molecular laboratories, they are unable to capture the sequence alterations across all exons of a gene and, for the same reason, have limited CNV detection capability. Consequently, NGS assays covering all exons are a substantial upgrade for screening patient tumours for mutations and CNVs.
In this study, we showed that the 409-gene NGS screen using the Ion Proton is an effective approach for a variety of somatic sequence variants (SNVs, insertions/deletions, and CNVs) in a wide variety of solid tumours with high concordance to other NGS platforms (IT-PGM and MiSeq) and Sanger sequencing. In addition, the higher sequencing capacity of the Ion Proton facilitated screening all exons of the genes, resulting in the detection of additional somatic variants. The Ion Proton (409-gene panel) also exhibited high sensitivity for most of the SNVs tested (up to 5% allelic fraction), had excellent inter-run and intra-run reproducibility, and used as little as 60 ng FFPE DNA, thereby rendering this approach well suited for routine diagnostic use.
The Ion Proton (409-gene panel) detected all expected SNVs in the samples with IT-PGM (46-gene panel) sequencing results unless there was amplicon failure in the 409-gene panel. The Ion Proton also detected and called several insertions and deletions that were not called by the IT-PGM (46-gene panel), which is attributable to the more recent and improved version of analysis software used on the Ion Proton (v3.6.2). However, it is interesting that the Ion Proton did not detect and call a 6-bp insertion in exon 20 of EGFR, a 6-bp insertion in exon 9 of KIT, and a 2-bp deletion in TP53. Visualisation of the Ion Proton sequencing reads in these genes showed these sequence aberrations, but they were missed by the software used (v3.6.2). Upon reanalysis of the data using a version recently released (v4.0.2), these variants were successfully detected. Reanalysis of an additional set of cell lines and tumours showed complete concordance of the software versions (Supplementary Table 4). An overall concordance of Ion Proton (409-gene panel) sequencing with orthogonal methods and concordance with the MiSeq (54-gene panel) sequencer established the accuracy and robustness of the Ion Proton (409-gene panel).
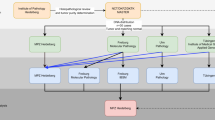
The high sequencing capacity of the Ion Proton permits the sequencing of all exons of 409 genes and ensures the detection of any mutation occurring in the entire coding region of the gene. This represents a major improvement and advantage over mutational hotspot screening panels using sequencers of lower capacity like the IT-PGM. This was evident in the detection of several additional somatic mutations by the Ion Proton (409-gene panel) in comparison to IT-PGM (46-gene panel). Of note, mutations in APC and PTEN were detected in gastrointestinal and breast carcinoma samples, respectively, that could not be detected using the 46-gene hotspot panel and the IT-PGM. Thus, the mutational status for these genes would be considered as wild-type if only hotspot panel-based testing were used for screening. The high sequencing capacity of the Ion Proton, which permits the simultaneous screening of all exons, facilitates comprehensive detection of all mutations in the tested genes and inspires more confidence in CNV detection. Furthermore, the capacity to simultaneously sequence up to 10 multiplexed samples per run render the 409-gene sequencing using the Ion Proton very suitable to provide timely mutational profiling of tumours with a turn-around-time of 5 days (workflow summarised in Supplementary Figure 4).
Overall, massive parallel sequencing of 409 genes using the Ion Proton to simultaneously detect SNVs, insertions, deletions, and gene CNVs was found to be a sensitive and comprehensive screen for somatic aberrations. Importantly, this assay required only a single expenditure of low-quantity FFPE DNA. The high level of reproducibility, sensitivity and FFPE-compatibility make the Ion Proton (409-gene panel) sequencing valuable for mutational screening of tumour samples in a clinical molecular diagnostics laboratory.
Change history
11 November 2014
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Beadling C, Neff TL, Heinrich MC, Rhodes K, Thornton M, Leamon J, Andersen M, Corless CL (2013) Combining highly multiplexed PCR with semiconductor-based sequencing for rapid cancer genotyping. J Mo Diagn 15 (2): 171–176.
Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, Ryu E, Targonski PV, Van Norstrand MD, Hathcock MA, Takahashi PY, McCormick JB, Johnson KJ, Maschke KJ, Rohrer Vitek CR, Ellingson MS, Wieben ED, Farrugia G, Morrisette JA, Kruckeberg KJ, Bruflat JK, Peterson LM, Blommel JH, Skierka JM, Ferber MJ, Black JL, Baudhuin LM, Klee EW, Ross JL, Veldhuizen TL, Schultz CG, Caraballo PJ, Freimuth RR, Chute CG, Kullo IJ (2014) Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc 89 (1): 25–33.
Bowcock AM (2013) Invited review DNA copy number changes as diagnostic tools for lung cancer. Thorax 69 (5): 495–496.
Dutton-Regester K, Hayward NK (2012) Whole genome and exome sequencing of melanoma: a step toward personalized targeted therapy. Adv Pharmacol 65: 399–435.
Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR (2008) The Catalogue of Somatic Mutations in Cancer (COSMIC). Curr Protoc Hum Gen 57: 10.11.1–10.11.26.
Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L (2012) A landscape of driver mutations in melanoma. Cell 150 (2): 251–263.
Li T, Kung HJ, Mack PC, Gandara DR (2013) Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol 31 (8): 1039–1049.
Liang H, Kim YH (2013) Identifying molecular drivers of gastric cancer through next-generation sequencing. Cancer Lett 340 (2): 241–246.
Luthra R, Patel KP, Reddy NG, Haghshenas V, Routbort MJ, Harmon MA, Barkoh BA, Kanagal-Shamanna R, Ravandi F, Cortes JE, Kantarjian HM, Medeiros LJ, Singh RR (2014) Next-generation sequencing-based multigene mutational screening for acute myeloid leukemia using MiSeq: applicability for diagnostics and disease monitoring. Haematologica 99 (3): 465–473.
Martelli MP, Sportoletti P, Tiacci E, Martelli MF, Falini B (2013) Mutational landscape of AML with normal cytogenetics: biological and clinical implications. Blood Rev 27 (1): 13–22.
Mwenifumbo JC, Marra MA (2013) Cancer genome-sequencing study design. Nat Rev Genet 14 (5): 321–332.
Newcombe RG (1998) Two-sided confidence intervals for the single proportion: comparison of seven methods. Stat Med 17 (8): 857–872.
Roberts NJ, Klein AP (2013) Genome-wide sequencing to identify the cause of hereditary cancer syndromes: with examples from familial pancreatic cancer. Cancer Lett 340 (2): 227–233.
Ross JS, Cronin M (2011) Whole cancer genome sequencing by next-generation methods. Am J Clin Pathol 136 (4): 527–539.
Ross JS, Wang K, Rand JV, Sheehan CE, Jennings TA, Al-Rohil RN, Otto GA, Curran JC, Palmer G, Downing SR, Yelensky R, Lipson D, Balasubramanian S, Garcia L, Mahoney K, Ali SM, Miller VA, Stephens PJ (2014) Comprehensive genomic profiling of relapsed and metastatic adenoid cystic carcinomas by next-generation sequencing reveals potential new routes to targeted therapies. Am J Surg Pathol 38 (2): 235–238.
Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J (2011) An integrated semiconductor device enabling non-optical genome sequencing. Nature 475 (7356): 348–352.
Singh RR, Patel KP, Routbort MJ, Reddy N, Barkoh B, Handal B, Kanagal-Shamanna R, Greaves WO, Medeiros LJ, Aldape K, Luthra R (2013) Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn 15 (5): 607–622.
Spencer DH, Sehn JK, Abel HJ, Watson MA, Pfeifer JD, Duncavage EJ (2013) Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. J Mol Diagn 15 (5): 623–633.
Thorvaldsdottir H, Robinson JT, Mesirov JP (2012) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14 (2): 178–192.
Tuna M, Amos CI (2013) Genomic sequencing in cancer. Cancer Lett 340 (2): 161–170.
Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, Baty J, Heath S, Westervelt P, Watson MA, Tomasson MH, Nagarajan R, O'Gara BP, Bloomfield CD, Mrozek K, Selzer RR, Richmond TA, Kitzman J, Geoghegan J, Eis PS, Maupin R, Fulton RS, McLellan M, Wilson RK, Mardis ER, Link DC, Graubert TA, DiPersio JF, Ley TJ (2009) Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci USA 106 (31): 12950–12955.
Workman P, Al-Lazikani B, Clarke PA (2013) Genome-based cancer therapeutics: targets, kinase drug resistance and future strategies for precision oncology. Curr Opin Pharmacol 13 (4): 486–496.
Acknowledgements
The authors are very thankful the patients and their families for the samples used in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Singh, R., Patel, K., Routbort, M. et al. Clinical massively parallel next-generation sequencing analysis of 409 cancer-related genes for mutations and copy number variations in solid tumours. Br J Cancer 111, 2014–2023 (2014). https://doi.org/10.1038/bjc.2014.518
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.518
Keywords
This article is cited by
-
Simplified molecular classification of lung adenocarcinomas based on EGFR, KRAS, and TP53 mutations
BMC Cancer (2020)
-
Comprehensive routine diagnostic screening to identify predictive mutations, gene amplifications, and microsatellite instability in FFPE tumor material
BMC Cancer (2020)
-
Integrated clinico-molecular profiling of appendiceal adenocarcinoma reveals a unique grade-driven entity distinct from colorectal cancer
British Journal of Cancer (2020)
-
Targeted capture-based NGS is superior to multiplex PCR-based NGS for hereditary BRCA1 and BRCA2 gene analysis in FFPE tumor samples
BMC Cancer (2019)
-
Comparison of somatic variant detection algorithms using Ion Torrent targeted deep sequencing data
BMC Medical Genomics (2019)