Abstract
Background:
Epigenetic therapy using histone deacetylase inhibitors (HDACi) has shown promise in clinical trials for the treatment of human malignancies. In addition to the immediate effects on the tumour cell growth, HDACi upregulates the expression of MHC class I-related chain molecules A and B (MICA and MICB), resulting in an enhanced susceptibility of tumour cells to natural killer cell-mediated lysis. The molecular mechanism underlying is still unclear.
Methods:
The transcriptional regulation mechanism underlying suberoylanilide hydroxamic acid (SAHA)-mediated regulation of MICA and related miRNA expression was investigated using promoter acetylation assays, bioinformatics analysis and chromatin immunoprecipitation assay.
Results:
SAHA upregulates the transcription of MICA/B by promoting MICA-associated histone acetylation while suppressing the MICA/B-targeting miRNAs miR-20a, miR-93 and miR-106b. The mechanism by which SAHA repressed miRNAs transcription involved repression of their host genes (miR-17-92 cluster and MCM7). SAHA downregulated the miR-17-92 cluster by abolishing tyrosine phosphorylation of STAT3 and decreased MCM7 transcription through localised histone deacetylation.
Conclusions:
The HDACi SAHA epigenetically upregulates MICA expression through regulating the expression of miR-17-92 cluster and MCM7 in hepatoma, thus enhancing the sensitivity of HCC to natural killer cell-mediated lysis. This novel mechanism of action provides promise for HDACi in therapy of HCC.
Similar content being viewed by others
Main
Epigenetic therapy using histone deacetylase inhibitors (HDACi) has shown promise in clinical trials for the treatment of human malignancies (Dawson and Kouzarides, 2012). Suberoylanilide hydroxamic acid (SAHA) has been approved by the Food and Drug Administration for the treatment of cutaneous T-cell lymphoma, and many other HDACi are undergoing clinical trials (Duvic et al, 2007; Sato, 2013). HDACi specifically induce cancer cell to differentiate, undergo cell cycle arrest and initiate apoptosis by altering the expression of genes involved in apoptosis signal transduction pathways or cell cycle modulation (Bolden et al, 2013). It is hypothesised that the accumulation of acetylated proteins, particularly histones, results in the upregulation of genes that have become epigenetically silenced (Khan and Khan, 2010). In particular, the gene encoding the cell cycle kinase inhibitor p21, is commonly upregulated in tumour cells treated with these agents (Richon et al, 2000; Yang et al, 2014). In addition to the immediate effects of HDACi on tumour cell growth, HDACi-induced recognition of tumour cells by immune cells also contributes to their anticancer effects (Setiadi et al, 2008). For example, we and others have reported that HDACi promote the expression of MICA or MICB, ligands of the natural killer (NK) cell activating receptor NKG2D on tumour cells, thus increasing their susceptibility to NK cell-mediated lysis (Armeanu et al, 2005; Zhang et al, 2009). The molecular mechanisms underlying the involvement of HDACi-regulated genes in immune recognition, however, are not fully understood. An increased understanding of the molecular mechanisms by which HDACi elicit immunostimulatory effects would undoubtedly contribute to their clinical development as anticancer agents.
MicroRNAs (miRNAs) are small, non-coding RNAs that can regulate various target genes. Computational and biological evidence suggests that miRNA-mediated gene regulation represents a fundamental mechanism of post-transcriptional regulation with diverse functional effects. Stern-Ginossar et al (2008) identified six cellular miRNAs, miR-20a, miR-93, miR-106b, miR-372, miR-373 and miR-520d, that target the mRNA of the MHC class I-related chain molecules A and B (MICA and MICB). Interestingly, among these miRNAs, both miR-106b and miR-93 are in the miR-106b-93-25 cluster while miR-20a is in the miR-17-92 cluster. Both miRNA clusters have been shown to accumulate in different types of cancer and have been designated oncomiRs, with miR-17-92 being designated oncomiR-1 (He et al, 2005). Maintenance complex component 7 (MCM7), the host gene of the miR-106b-93-25 cluster, is also observed to have a higher expression level in many cancers and is regarded as an indicator of poor prognosis (Poliseno et al, 2010). The identification of miRNA target genes has been extensively pursued, whereas little is known about the mechanism of regulation of miRNA by epigenetic alterations. MICA/B-targeting miRNAs are overexpressed in tumours, and were found to promote tumour progression and contribute to avoidance of immune recognition via their suppression of MICA and MICB expression.
Early models of regulation of transcription by acetylation focused on the physical interactions of basic histone proteins with negatively charged DNA. The addition of an acetyl group is believed to neutralise the positive charge of histones, decreasing their interactions with negatively charged DNA. This results in de-compaction of chromatin and greater access to DNA for transcription factors, leading to a transcriptionally active genomic locus. There is, however, considerable evidence that these models are oversimplified (Drummond et al, 2005). Not all genes are upregulated by HDACi treatment. In fact, the ratio of upregulated to downregulated genes is close to 1 : 1. This suggests that the regulation of gene expression by acetylation is highly selective and also likely involves chromatin-associated non-histone proteins. Additionally, HDACi can downregulate certain oncogenes (Kim et al, 2001; Mitsiades et al, 2004; Chou et al, 2011) via reduced transcription because of local histone hypoacetylation after HDAC and HAT dissociation from the respective promoter regions (Duan et al, 2005; Fiegler et al, 2013).
In this present study, we investigated the molecular mechanisms underlying SAHA-mediated regulation of MICA and miRNA expression in two hepatocellular carcinoma (HCC) cell lines. The results demonstrated that SAHA significantly increased MICA/B levels by promoting localised histone acetylation, and simultaneously suppressed the miRNAs targeting MICA/B by localised histone deacetylation or by abolishing tyrosine phosphorylation of STAT3. Both mechanisms likely contribute to SAHA—induced increases in the expression of MICA and MICB proteins. Expression of MICA/B enhances immune recognition of tumours by innate immune cells, and ultimately enhances their sensitivity to cytolysis by NK cells.
Materials and methods
Cell culture
The human HCC cell lines HepG2 (The Cell Bank of Type Culture Collection of Chinese Academy of Sciences) and H7402 (Institute of Basic Medical Sciences, Shandong Academy of Medical Science, Jinan, China) were cultured in RPMI-1640 medium (GIBCO/BRL, Grand Island, NY, USA) containing 10% foetal bovine serum. The human NK cell line NKL, generously provided by Dr Jin Boquan (Fourth Military Medical University, Xi'an, China), were cultured in RPMI-1640 medium containing 10% foetal bovine serum and 100 U ml−1 rhIL-2 (Changsheng, Changchun, China). All cells were incubated at 37 °C in a 5% CO2 atmosphere and passaged once every 2–3 days. Experiments were initiated when cells showed logarithmic growth.
Reagents
SAHA, which was synthesised by Department of Medicinal Chemistry, School of Pharmacy, Shandong University, Shandong, China, was stored at −20 °C, dissolved in DMSO to a 5 mM stock, and diluted with culture medium as needed. IL-6, a kind gift of Professor Rui Sun (Department of Microbiology and Immunology, School of Life Sciences, University of Science and Technology of China, Hefei, China), was stored at 4 °C and dissolved as a 1 × 107 U ml−1 stock in 1 × phosphate buffered saline.
Cytotoxicity assay
After treatment with 2 μ M SAHA for 48 h, HepG2 and H7402 cells were used as target cells and plated in 96-well plates at a density of 1.0 × 104 cells per well. NK cells were coincubated at an effector-to-target ratio between 4 : 1 and 1 : 1 for 6 h. Subsequently, 20 μl methylthiazolyldiphenyl-tetrazolium bromide (5 mg ml−1) was added to each well and incubated for an additional 4 h (Qu et al, 2014). Absorbance at 570 nm and 630 nm was measured using a microplate reader (Synergy 2, BioTek, Winooski, VT, USA). Cytotoxicity was calculated using the following formula: cytotoxicity (%)=1−(ODE+T−ODE)/ODT × 100% (ODE+T: OD value of the effector cell and target cell group; ODE: OD value of the effector cell group; ODT: OD value of the target cell group).
Flow cytometry
For cell surface protein expression analysis, cells were harvested and washed with phosphate buffered saline, and incubated with antibodies for 1 h at room temperature. The following antibodies were used: anti-MICA/B mAb (eBioscience, San Diego, CA, USA); anti-MICA mAb (R&D Systems, Minneapolis, MN, USA); and anti-HLA-A/B/C mAb (BD Pharmingen, San Diego, CA, USA). Measurements were performed using a flow cytometer (FACScalibur; BD Biosciences, San Jose, CA, USA) and analysed with WinMDI 2.9 software (Scripps Institute, La Jolla, CA, USA).
RNA isolation and quantitative reverse transcription-PCR analysis of mRNA and miRNA
RNA was extracted using a TRIzol RNA isolation kit (Invitrogen, Carlsbad, CA, USA). MiR-20a, miR-93, miR-106b, miR-17, miR-18a, miR-19a were quantified by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) using specific BulgeLoop miRNA qRT-PCR primers purchased from Guangzhou Ribobio (Guangzhou, China) with U6 small nuclear RNA as an internal reference (Hou et al, 2014). The mRNA expression levels of pri-miR-17-92 and MCM7 were determined by qRT-PCR. Expression levels of the target genes were normalised to GAPDH. The sequences of the PCR primers are listed in Supplementary Tables 1 and 2. Real-time PCR was carried out using SYBR green mix (FastStart Universal SYBR Green Master, Roche, Indianapolis, IN, USA).
miRNA mimics transfection
MiRNA mimics (double-stranded oligonucleotides) used for the overexpression of miR-20a, miR-93 and miR-106b in hepatoma cells were purchased from GenePharma (Shanghai, China). H7402 or HepG2 cells were seeded at a density of 1.5 × 105 cells ml−1, and transfected with miR-20a/miR-93/miR-106b mimics with Lipofectamine2000 (Invitrogen) according to the manufacturer’s instructions. The miRNA mimics were used at final concentration of 100 nM. Negative controls of miRNA mimics (mNC) were transfected as matched controls (Su et al, 2011).
Chromatin immunoprecipitation (ChIP) assay
H7402 cells and HepG2 cells were plated at a density of 1 × 106 cells in 10-cm dishes and cultured with 2 μ M SAHA for 48 h. Chromatin immunoprecipitation (ChIP) was performed using the EZ ChIP kit (Millipore, Temecula, CA, USA) according to the manufacturer's recommendations. Briefly, cells were lysed and crosslinked with 1% formaldehyde. Lysates were sonicated to shear DNA to lengths between 200 and 1000 base pairs. An aliquot of the chromatin preparation was set aside and designated as the input fraction. Immunoprecipitations were carried out in ChIP Dilution Buffer with the following antibodies: IgG, anti-acetyl histone H3 (AcH3), anti-acetyl histone H4 (AcH4; Millipore), and anti-p-Tyr705-STAT3 (Cell Signaling Technology, Danvers, MA, USA). Immunoprecipitates were collected using protein G-sepharose beads, washed and eluted according to the manufacturer’s protocol. Immunoprecipitated DNA was recovered by reversing crosslinking, and purified using the QIAGEN Purification Kit (QIAGEN, Hilden, Germany) and analysed by PCR. The sequences of the PCR primers used for MICA, miR-17-92 or MCM7 promoter analysis are listed in supplementary Table 3. Negative control primers (Millipore) amplified a non-related GAPDH promoter sequence.
Western blot
H7402 cells and HepG2 cells were treated with SAHA (1-2 μ M) for 48 h. Western blotting assays were carried out as described previously (Zhang et al, 2014). The following antibodies were used: anti-p-Ser727-STAT3, anti-MCM7 mAb (BBI; Shanghai SangonBiotech, CHN); anti-STAT3, anti-p-Tyr705-STAT3 (Cell Signaling Technology); AcH3 mAb, and AcH4 mAb.
Statistical analysis
Significant differences were determined by Student's t-test and one way-ANOVA. P<0.05 was considered significant. Data are presented as the mean±s.d. of three independent experiments. All analyses were performed with GraphPad Prism 5 (La Jolla, CA, USA).
Results
SAHA treatment significantly increased the susceptibility of HCC cell lines to cytolysis by NK cells
In many cases, the functions of NK cells in cancer patients are impaired and tumours can escape NK cell-mediated cytotoxicity. The HCC cell lines HepG2 and H7402 were treated with 2 μ M SAHA, a concentration that does not induce significant cell apoptosis (data not shown) for 48 h. Sensitivity of these cells to NK-mediated lysis was then evaluated. The results showed that SAHA treatment significantly increased the susceptibility of the two HCC cell lines to cytolysis by NKL cells (Figure 1A). Furthermore, we found that the expression of MICA/B was upregulated, while there were no significant changes in expression of HLA-ABC in the two HCC cell lines when exposed to SAHA (Figure 1B). These results suggest that SAHA treatment may increase expression of MICA/B in hepatoma cells, and further promote the interaction between NKG2D and its ligands, MICA and MICB, thus increasing the susceptibility of HCC cells to cytotoxic NK cells.
SAHA sensitises HCC cells to IL-2–activated NK cell-mediated cytotoxicity by upregulating MICA. (A) Human HCC cell lines H7402 and HepG2 were untreated (Ctrl) or treated with SAHA (2 μ M). After 48 h, cells were coincubated with NKL cells for 6 h, and the cytotoxicity of NK cells was measured in the MTT assay. (B) HCCs cells were treated with SAHA. After 48 h, the expression of HLA-ABC, MICA/B and MICA was analysed by flow cytometry. Shown is one representative experiment. The data represent the mean ±s.d. of three independent experiments. *P< 0.05, vs Ctrl group.
Expression of miRNAs targeting MICA/B is downregulated in SAHA-treated HCC cells in a dose-dependent manner
MiR-20a, miR-93, miR-106b, miR-372, miR-373 and miR-520d regulate the expression of MICA and MICB (Stern-Ginossar et al, 2008), whereas miR-20a, miR-93 and miR-106b have been implicated in tumourigenesis. To explore the molecular mechanisms of SAHA in the regulation of MICA/B expression, we sought to examine whether SAHA could regulate the expression of these miRNAs. Among the panel of miRNAs, only miR-20a, miR-93 and miR-106b were detectable in the cell lines tested. SAHA treatment resulted in a two- to four-fold dose-dependent decrease in the expression of miR-20a and miR-106b in both HCC cell lines (Figure 2). MiR-93 also showed a modest decrease in HepG2 cells. Downregulation of these miRNAs was consistent with upregulation of MICA/B, suggesting that SAHA increased MICA/B expression through downregulation of these MICA/B-targeting miRNAs, further enhancing susceptibility of HCC cells to NK cell-mediated lysis.
Expression of miRNAs targeting MICA is downregulated in a dose-dependent manner in SAHA-treated HCC cell lines. qRT-PCR analysis was performed of miR-20a, miR-93, and miR-106a in H7402 and HepG2 cells treated with varying concentrations of SAHA (0.5, 1, 2, 4 μ M) for 48 h. Relative expression levels were normalised to U6 gene expression levels. Error bars represent the s.d. of mean values (expressed as percent of control) from three independent experiments. **P<0.01; *P<0.05, paired t-test.
Expression of miR-17-92 cluster and MCM7, the host genes of miRNA targeting MICA/B, is downregulated in SAHA-treated HCC cells in a dose-dependent manner
Approximately 500–1000 miRNAs are expressed in human cells, and their expression signatures vary depending on the tissue and cell type examined (O'Connell et al, 2012). MiRNAs are transcribed by RNA polymerase II as long primary transcripts (pri-miRNAs) that undergo sequential processing to produce mature miRNAs (Lee et al, 2004). There are at least two types of miRNA promoters. Some miRNAs have their own promoters, while others are located within an mRNA or intron and are transcribed as part of the host gene (Takada and Asahara, 2012). MiR-17-92, which is processed from the transcript of Chromosome 13 open reading frame 25 (C13orf25), a target gene for 13q31–q32 amplification (Ota et al, 2004) in some lymphomas and solid tumours (Ji et al, 2011), is the host gene of miR-20a. In addition to miR-20a, miR-17, miR-18a and miR-19a are also memebers of miR-17-92 cluster and located at the upstream of miR-20a. Minichromosome MCM7 is the host gene of miR-93 and miR-106b. To address whether SAHA might repress miR-20a, miR-93 and miR-106b transcription by suppressing the expression of their host genes, we examined the expression of MCM7 and miR-17, miR-18a, miR-19a in the miR-17-92 cluster. Primers for qPCR to detect pri-miRNA transcripts of miR-17-92 were also designed (Nagel et al, 2009; Yan et al, 2009). When HCC cells were treated with SAHA (0.5, 1, 2, 4 μ M) for 24 and 48 h, dose- and time-dependent decreases in the levels of miR-17, miR-18a and miR-19a were observed (Figure 3A). Interestingly, they were repressed to the same extent as miR-20a was repressed (Figure 2). Furthermore, the levels of pri-miR-17-92 and MCM7 also decreased significantly in both a dose- and time-dependent manner (Figure 3B and C). Together, these results demonstrate that treatment with SAHA results in reduced expression of the host genes of miRNA targeting MICA/B miR-17-92 cluster and MCM7 in the two HCC cell lines.
Expression of miR-17, miR-18a and miR-19a in C13ORF25 BOX and MCM7 is downregulated in a dose-dependent manner in SAHA-treated HCC cell lines. (A) qRT-PCR analysis of miR-17, miR-18a, miR-19a and (B) the primary miRNA transcript (pri-miR-17-92) of their host gene miR-17-92 expression in H7402 and HepG2 cells treated with SAHA at different concentrations (0.5, 1, 2, 4 μ M) for 24 h and 48 h.(C) qRT-PCR analysis of MCM7 (the host gene of miR-93, miR-106b) expression in H7402 cells.
miR-20a, miR-93 and miR-106b specifically downregulate MICA expression in HCC cell lines
We next examined whether these miRNAs could affect MICA surface protein levels. MiRNA mimics or control miRNAs were transiently transfected into HepG2 or H7402 cells, which had high expression of both MICA and MICB. As shown in Figure 4, all three miRNAs significantly downregulated MICA surface expression. Our findings are consistent with previous studies from Stern-Ginossar, which showed that miR-20a, miR-93 and miR-106b target the 3′ untranslated region of MICA and MICB, and suppress MICA/B expression (Stern-Ginossar et al, 2008). These data suggest a novel molecular mechanism by which HDACi regulate MICA expression. HDACi may promote the expression of the NKG2D ligand MICA by downregulating the expression of miRNAs (miR-20a, miR-93 and miR-106b) targeting MICA/B.
MiR-20a, miR-93 and miR-106b specifically downregulate MICA expression in HCC cell lines. H7402 cells and HepG2 cells were transfected with 100 nM of the indicated human miRNA (red lines) or control miRNA (black lines) or untransfected (dotted lines). After 24 h, MICA expression was analysed by flow cytometry. Filled histograms represent staining with a control antibody. Shown is a representative trace selected from at least three independent experiments.
SAHA induces the accumulation of acetylated histones in chromatin associated with the MICA gene, but decreases the binding of AcH4 to the MCM7 promoter
To verify the activity of the HDACi in our studies, HepG2 cells were treated with SAHA for 48 h, and AcH3 and AcH4 protein levels were determined by western blot analysis. As shown in Figure 5A, neither AcH3 nor AcH4 were detectable in HepG2 cells treated with the vehicle (DMSO) control. Following treatment with SAHA, the levels of AcH3 and AcH4 were increased in a dose-dependent manner (P<0.05).
SAHA induces accumulation of acetylated histones in chromatin associated with the MICA gene, but decreases the binding of AcH4 to the MCM7 promoter. H7402 cells and HepG2 cells were treated with SAHA (1-2 μ M) for 48 h. (A) Western blot analysis showed that SAHA induced an increase in AcH3 and AcH4 in HepG2 cells. (B) Schematic representation of the human MICA gene. Primer sets are indicated as P1, P2, and P3. (D) Schematic representation of the human miR-17-92 gene. Primer sets are indicated as AmiR, BmiR, CmiR, DmiR. (F) Schematic representation of the human MCM7 gene. Primer sets are indicated as PM1, PM2.(C, E, G) Soluble chromatin was immunoprecipitated with indicated antibodies. PCR primers for the regions of the MICA gene, miR-17-92 gene, MCM7 gene as indicated above were used to amplify the DNA isolated from the immunoprecipitated chromatin. Experiments were carried out at least three times.
As demonstrated above, SAHA induces MICA/B expression in HCC cells when compared with vehicle (DMSO) control (P<0.05; Figure 1B). To determine whether these changes were associated with histone acetylation status in the promoter region of MICA, ChIP was performed using primers corresponding to -1105 (P1), -439 (P2) and -308 (P3) bp upstream of the translation initiation site (ATG) of the MICA gene (Figure 5B). Chromatin fragments from HCC cells cultured with 2 μ M SAHA were immunoprecipitated with antibody to AcH3 or AcH4. DNA from the immunoprecipitation was isolated, and a 213-bp fragment of the MICA promoter region was amplified (Figure 5C). Following culturing with SAHA for 48 h, a 3.7-fold increase in MICA promoter DNA in H7402 cells and 3.2-fold increase in MICA promoter DNA in HepG2 cells was associated with highly AcH3, compared with the same region isolated from HCC control cells. PCR analysis with the P2 primer, amplifying a 423-bp fragment of the MICA promoter region and the P3 primer, amplifying a 310-bp fragment of the MICA promoter region was also performed (Figure 5C). The accumulation of AcH4 in chromatin associated with the MICA gene was verified in HepG2 cells (Figure 5C). These results strongly suggest that the enhancement in acetylation of histones associated with the MICA promoter is involved in the upregulation of MICA induced by SAHA.
We also examined histone modification in the promoter region of miR-17-92. ChIP primers were designed corresponding to -4913 (A), -4181 (B) (Woods et al, 2007), -4003 (C) and -3927 (D) bp upstream of the translation initiation site of the miR-17-92 gene (Figure 5D). No increases in the levels of AcH3 and AcH4 were found at the promoter region of miR-17-92 in HCC cells treated with SAHA (Figure 5E). We further performed quantitative ChIP assays to examine the binding of acetylated histones to the MCM7 promoter regions. Primers were designed at -2273 (PM1) and -2194 (PM2) base pairs relative to the transcription initiation site of the MCM7 gene (Figure 5F) (Scian et al, 2008; Zhao et al, 2012). Consistent with SAHA-induced MCM7 transcriptional repression, ChIP analysis showed a significant reduction of AcH4 binding to both MCM7 promoters, although SAHA treatment did not alter binding of AcH3 to either promoter region (Figure 5G). This demonstrates the selectivity of gene regulation by acetylation. The observed histone H4 hypoacetylation could be the result of reduced cooperative HDAC/HAT binding and activity at the MCM7 5′-UTR, even though SAHA strongly enhances global histone acetylation levels. Taken together, these data suggest that HDACi may regulate the transcription of MICA by modifying histone acetylation, and regulate the transcription of MCM7 by a localised decrease of histone H4 acetylation at its promoters. Histone acetylation did not affect transcription of the miR-17-92 cluster directly during SAHA treatment.
SAHA downregulated the miR-17-92 cluster by abolishing tyrosine phosphorylation of STAT3
To investigate the mechanisms of downregulation of the miR-17-92 cluster by SAHA, we analysed the sequences of the miR-17-92 cluster to predict the exact location of its promoter with Promoter 2.0. The binding sites of transcription factors in the promoter regions of miR-17-92 were predicted using the JASPAR database and TFSEARCH. The promoter regions of miR-17-92 were determined to contain putative transcription factor binding sites, such as, GATA-1, GATA2, GATA-3, SP1, E2F, c-myc and STAT3. Previous studies have demonstrated that both E2F and c-myc can transcriptionally activate the miR-17-92 cluster (O'Donnell et al, 2005; Petrocca et al, 2008). We found that expression of SP1 was unchanged following SAHA treatment (data not shown). GATA-3, an essential regulator of T-lymphocyte differentiation (Ting et al, 1996), does not appear to play a role in the development of hepatic carcinoma. We, therefore, focused on the tumour-associated transcription factors GATA-2 and STAT3 (Luesink et al, 2012). Although the protein levels of GATA-2 were decreased after SAHA treatment (Supplementary Figure 1A), overexpression of GATA-2 (Supplementary Figure 1B, 1C) had no effect on the expression of miR-17-92 (Supplementary Figure 1D). STAT3 has been reported to transcriptionally activate the miR-17-92 cluster and paralogous clusters (Brock et al, 2009). Increasing evidence suggests that the regulation of gene expression by STATs requires class I HDAC activity (Gupta et al, 2012). Therefore, we next attempted to determine whether SAHA treatment had any effect on the expression or activation of STAT3 in HCC cells. As shown in Figure 6A, both H7402 and HepG2 cells showed high levels of phosphorylation at tyrosine 705 of STAT3. SAHA treatment resulted in the dephosphorylation of STAT3 in both HCC cell lines within 48 h with little effect on total STAT3 levels (Figure 6A). Similar results were obtained with regards to phosphorylation of STAT3 at serine 727 (Figure 6B). We also observed decreases in the protein levels of MCM7, the host gene of miR-93 and miR-106b, in response to SAHA treatment in whole cell lysates (Figure 6B).
SAHA downregulates the miR-17-92 cluster by abolishing phosphorylation of STAT3. (A, B) H7402 cells and HepG2 cells were treated with SAHA (1-2 μ M) for 48 h. Cell lysates were subjected to Western blot analysis with anti-STAT3, anti-p-STAT3 (Tyr705), anti-p-STAT3 (Ser727) and anti-MCM7 antibodies. (C) The levels of STAT3 and MCM7 in H7402 cells stimulated with IL-6 were examined by Western blotting. (D) The levels of p-STAT3 (Tyr705) and p-STAT3 (Ser727) in H7402 cells and HepG2 cells stimulated with IL-6 for indicated times were examined by Western blotting. (E) qRT-PCR analysis of miR-20a expression in H7402 and HepG2 cells treated with IL-6. (F) qRT-PCR analysis of pri-miR-17-92 expression in H7402 cells treated with IL-6. (G, H) H7402 cells and HepG2 cells were stimulated with IL-6 alone or with IL-6 combined with 2 μ M SAHA for 24 h. The levels of p-STAT3 (Tyr705) and AcH3 protein were examined by Western blotting. MiR-20a expression in H7402 and HepG2 cells was detected by qRT-PCR. *P<0.05, vs vehicle control group. #P<0.05, vs IL-6 group. Data shown are mean±s.d. of at least three independent experiments.
IL-6 is a known inducer of the STAT3 pathway. We stimulated the two HCC cell lines with IL-6 to induce the phosphorylation of STAT3. As shown in Figure 6C and D, higher levels of total STAT3 and phosphorylation of STAT3 was observed at 30 min, and sustained for at least 1 h. After stimulation with IL-6 for 24 h, the expression of miR-20a and pri-miR-17-92 increased significantly in both cell lines (Figure 6E and F). A relationship between activation of STAT3 and induction of miR-17-92 expression is possible. Importantly, SAHA treatment alleviated IL-6-induced STAT3 activation and the corresponding upregulation of miR-20a expression (Figure 6G and H).
Transcription Element Search System-mediated (JASPAR database) sequence analysis was performed and revealed three putative STAT3 sites scattered within the promoter region of the miR-17-92 cluster (Figure 7A). To determine whether activated STAT3 could directly bind to the promoter region of the miR-17-92 cluster and regulate transcription, ChIP analysis was performed in H7402 cells to validate promoter binding using an antibody to p-STAT3(Tyr705) followed by PCR using the primers covering the putative STAT3 binding sites. The results showed that the -4234 and -3709 sites had STAT3 occupancy in H7402 cells, while no binding of STAT3 was found at the -3317 site. As shown in Figure 7B, we verified that p-STAT3 could bind to the miR-17-92 promoter, with a reduction in STAT3 levels at the miR-17-92 promoter after SAHA treatment. We concluded that STAT3 promotes the expression of the miR-17-92 cluster, while SAHA treatment downregulates the miR-17-92 cluster via suppression of STAT3 phosphorylation.
SAHA supresses the binding of activated STAT3 to the promoter region of the miR-17-92 cluster. (A) Schematic representation of the human miR-17-92 gene. Primer sets are indicated as PI, PII, PIII, occupying predicted STAT3 binding sites within miR-17-92 promoter. (B) H7402 cells were treated with 2 μ M SAHA for 48 h. A ChIP assay was performed using anti-p-Tyr705-STAT3 antibody or IgG antibody as a negative control.
Discussion
HCC cells display a marked resistance to conventional cytostatic agents resulting in disappointing clinical outcomes when currently available chemotherapeutic treatment strategies are employed. There is growing evidence that HDAC inhibitors might improve the efficacy of standard chemotherapy. However, the molecular mechanisms underlying the anticancer effects of these drugs are not fully understood. Improved understanding of the molecular mechanisms would undoubtedly assist their clinical development as anticancer agents. The functions of NK cells are usually impaired in cancer patients. Manipulation of NK cell activation, therefore, has been regarded as an important approach to cancer immunotherapy. NKG2D is a pivotal activation receptor for NK cells. We and others have shown that treatment with HDACi induces expression of the NKG2D ligands MICA/B on some tumour cells, rendering them more sensitive to NK cell-mediated killing (Armeanu et al, 2005; Zhang et al, 2009). The present study further explores the molecular mechanism of regulation of MICA/B expression by the histone deacetylase inhibitor SAHA from the perspective of epigenetics and miRNA regulation. Our results demonstrate that HDACi selectively regulate gene expression, with increased histone acetylation near the MICA promoter and inhibition of the transcription of MICA/B-targeting miRNAs.
Histone acetylation status is assumed to be an important factor that controls the accessibility of transcription factors to DNA and subsequent gene transcription. Both AcH3 and AcH4 are associated with an open chromatin structure and active gene expression. For example, the p21WAF1 protein, an inhibitor of cyclin-dependent kinases, stalls cell cycle progression in the G1 phase. The induction of p21WAF1 by HDACi is associated with the accumulation of acetylated histones at the promoter region of the gene (Yang et al, 2014). In the present study, we demonstrated that the significant upregulation of MICA induced by SAHA was mainly a result of enhanced acetylation of histones H3 and H4 associated with the MICA promoter. Interestingly, the miR-17-92 cluster and its related paralogues (miR-106a/363 and miR-106b/25) showed decreased expression rather than activation. Although the mechanism of HDACi in the upregulation of gene expression in transformed cells has been extensively studied, the mechanism of downregulation of gene expression by HDACi is not fully understood. It has been reported that HDACi can suppress the expression of target genes, such as cyclin D1 (Alao et al, 2006), cyclinB1 (Mateen et al, 2012), bcr-abl (Nimmanapalli et al, 2003), ErbB2 (Drummond et al, 2005) and steroidogenic factor 1 (Chen et al, 2007), through proteasomal degradation or ubiquitin-dependent protein degradation at the protein level, whereas the repression of miRNAs by HDAC inhibitors occurs at the level of transcription. Understanding the regulatory mechanism of HDACi on miRNA expression is challenging.
We analysed the sequence of the miR-17-92 cluster to predict the exact location of its promoter using Promoter 2.0. The results revealed that the promoter of miR-17-92 cluster is most likely located approximately -4300 upstream of the transcription initiation site (ATG) of the miR-17-92 cluster. The sequences of predicted miR-17-92 promoters were then analysed for potential transcription factor binding sites using the JASPAR database and TFSEARCH. We found potential binding sites for GATA2 and STAT3 in the upstream promoter region of the miR-17-92 gene. SAHA suppressed the expression of GATA-2 in a dose-dependent manner, especially in K562 cells (Supplementary Figure 1). Overexpression of GATA-2 led to no changes in the expression of the mature transcripts of miR-20a, suggesting that GATA-2 may not be involved in the repression of miR-17-92 expression induced by SAHA.
Previous reports have suggested that STAT3 transcriptionally activates the miR-17-92 cluster (Brock et al, 2009). There is little direct evidence, however, regarding its precise regulatory mechanism. As an oncogene, STAT3 is persistently activated in many human cancers and transformed cell lines. After activation, STAT3 proteins dimerise and translocate to the nucleus, where they bind to specific DNA-response elements in the promoters of target genes and regulate gene expression. STAT3 was found in both the cytoplasm and nucleus, whereas activated p-STAT3 (Tyr705) was restricted to the nucleus. Our previous studies have demonstrated that STAT3 is constitutively activated in human HCC cell lines, and blockage of STAT3 activation suppresses growth of cancer cells, promotes cancer cell apoptosis and inhibits cell cycle progression in vitro (Sun et al, 2008). It is important to note that the activity of STAT3 can be post-translationally modulated by acetylation and phosphorylation, and both processes are affected by HDAC inhibitors. Gupta reported that treating cells with the HDACi LBH589 (panobinostat) increases STAT3 acetylation and results in decreased levels of STAT3 in the nucleus along with a significant decrease in nuclear p-STAT3 (Tyr). These results suggest that HDAC inhibition may enhance STAT3 export from the nucleus to the cytoplasm or prevent entrance of STAT3 to the nucleus (Gupta et al, 2012). Here, we demonstrated that SAHA treatment resulted in the dephosphorylation of STAT3 in a dose-dependent manner in HCC cells. We further demonstrated that p-STAT3 could directly bind to the miR-17-92 cluster promoter, with a reduction of p-STAT3 at the miR-17-92 promoter after SAHA treatment. Although HDAC inhibitors increase histone acetylation in treated cells, no increases occur at the miR-17-92 promoter region. Combined with decreased binding of p-STAT3 at the promoter, this results in the transcriptional repression of miR-17-92 by HDAC inhibitors. Besides, c-Myc (O'Donnell et al, 2005) and E2F1 (Petrocca et al, 2008) have been shown to transcriptionally activate the miR-17-92 cluster and paralogous clusters, while p53 (Yan et al, 2009) represses them (Grillari et al, 2010). Consistent with these reports, we verified that SAHA decreased the levels of E2F1 and c-myc in two HCC cell lines (data not shown). C-Myc and E2F1 may also be involved in the transcriptional repression of miR-17-92 in SAHA treated HCC cells.
The miR-106b-93-25 cluster is composed of the highly conserved miR-106b, miR-93 and miR-25, which have been shown to accumulate in different types of cancer. MCM7, the host gene of the miR-106b-93-25 cluster, is a transcription factor. High expression levels of MCM7 are also observed in many cancers and are regarded as an indicator of poor prognosis. In this study, we found that SAHA treatment significantly suppressed expression of the miR-106b-93-25 cluster as well as its host gene MCM7 in HCC cells. We further demonstrated that the transcriptional repression of the miR-106b-93-25 cluster and MCM7 by SAHA was associated with deacetylation of histone H4 but not H3 localised at the MCM7 promoter. Consistent with these findings, similar mechanisms have also been proposed for HDACi-induced regulation of expression of other genes. Studies by Ferguson (Ferguson et al, 2003) suggested that HDAC inhibitors could induce localised promoter histone deacetylation. Studies by Duan (Duan et al, 2005) showed that, although HDACi increased the global accumulation of acetylated histones, decreased AcH3 binding to the bcl-2 promoter regions was correlated with the transcriptional repression of bcl-2, while AcH4 binding did not change significantly. Noh also reported TSA (trichostatin A) augmented acetylation of the p21WAF1 promoter but reduced acetylation of the cyclin B1 promoter (Noh and Lee, 2003), suggesting a relationship between TSA-induced modulation of histone acetylation and differential expression of these genes. Fiegler (Fiegler et al, 2013) reported that HDACi downregulated B7-H6 through decreased B7-H6 promoter activity and reduced histone acetylation at the B7-H6 promoter. Local histone hypoacetylation induced by HDACi affects transcription by preventing recruitment of RNA polymerase II (You et al, 2008), as well as by reducing the binding of transcription factors (Duan et al, 2005). These observations are surprising because, in striking contrast to the usual correlation, histone acetylation is associated with deceased transcriptional activity (Struhl, 1998). The contribution of individual HATs to modulation of MCM7 expression requires further investigation. Although Li (Li et al, 2013) reported STAT3 may regulate the expression of MCM7 in mouse, no direct evidence was provided. We also attempted to determine whether STAT3 was involved in the regulation of MCM7 expression. We performed human MCM7 promoter analysis for transcription factor binding sites, and the result revealed several putative STAT3 sites scattered within the promoter region of MCM7 (data not shown). However, as shown in Figure 6C, although IL-6 stimulated the activation of STAT3 in H7402 cells, the expression of MCM7 was not significantly changed. These results suggested that STAT3 activation had no effect on the expression of MCM7 in HCC cells. In future studies, it will be important to perform a detailed characterisation of the MCM7 promoter to identify the transcription factors that are involved in the regulation of MCM7 expression by HDACi.
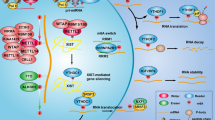
Based on our results, we propose a model to explain the role of SAHA in the regulation of gene expression in HCC cells (Supplementary Figure 2). On the one hand, SAHA induces MICA mRNA transcription through increases in MICA-associated histone acetylation; on the other hand, SAHA suppresses the miRNAs targeting MICA/B to reduce the threshold for MICA and MICB expression. Both mechanisms could contribute to the SAHA–induced increase in the expression of MICA and MICB protein, which promotes immune recognition of tumours by innate immune cells. We demonstrated that the influence of HDACi on transcriptional activity is not only dependent on primary histone targets but also on targeting non-histone protein. SAHA repressed the transcription of miR-20a, miR-93 and miR-106b by repressing their host genes (miR-17-92 cluster and MCM7). SAHA decreased miR-17-92 transcription by decreasing phosphorylation of tyrosine residues in STAT3, and reduced MCM7 mRNA transcription through localised promoter histone deacetylation. MiRNAs can, therefore, alter the epigenetic machinery and also be regulated by epigenetic alterations. This suggests that HDAC inhibitors may have therapeutic potential for patients with tumours that overexpress pSTAT3, MCM7, as well as the miR-17-92 cluster.
Change history
06 January 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Alao JP, Stavropoulou AV, Lam EW, Coombes RC, Vigushin DM (2006) Histone deacetylase inhibitor, trichostatin A induces ubiquitin-dependent cyclin D1 degradation in MCF-7 breast cancer cells. Mol Cancer 5: 8.
Armeanu S, Bitzer M, Lauer UM, Venturelli S, Pathil A, Krusch M, Kaiser S, Jobst J, Smirnow I, Wagner A, Steinle A, Salih HR (2005) Natural killer cell-mediated lysis of hepatoma cells via specific induction of NKG2D ligands by the histone deacetylase inhibitor sodium valproate. Cancer Res 65 (14): 6321–6329.
Bolden JE, Shi W, Jankowski K, Kan CY, Cluse L, Martin BP, MacKenzie KL, Smyth GK, Johnstone RW (2013) HDAC inhibitors induce tumor-cell-selective pro-apoptotic transcriptional responses. Cell Death Dis 4: e519.
Brock M, Trenkmann M, Gay RE, Michel BA, Gay S, Fischler M, Ulrich S, Speich R, Huber LC (2009) Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ Res 104 (10): 1184–1191.
Chen WY, Weng JH, Huang CC, Chung BC (2007) Histone deacetylase inhibitors reduce steroidogenesis through SCF-mediated ubiquitination and degradation of steroidogenic factor 1 (NR5A1). Mol Cell Biol 27 (20): 7284–7290.
Chou CW, Wu MS, Huang WC, Chen CC (2011) HDAC inhibition decreases the expression of EGFR in colorectal cancer cells. PLoS One 6 (3): e18087.
Dawson MA, Kouzarides T (2012) Cancer epigenetics: from mechanism to therapy. Cell 150 (1): 12–27.
Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC (2005) Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol 45: 495–528.
Duan H, Heckman CA, Boxer LM (2005) Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol Cell Biol 25 (5): 1608–1619.
Duvic M, Talpur R, Ni X, Zhang C, Hazarika P, Kelly C, Chiao JH, Reilly JF, Ricker JL, Richon VM, Frankel SR (2007) Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL). Blood 109 (1): 31–39.
Ferguson M, Henry PA, Currie RA (2003) Histone deacetylase inhibition is associated with transcriptional repression of the Hmga2 gene. Nucleic Acids Res 31 (12): 3123–3133.
Fiegler N, Textor S, Arnold A, Rolle A, Oehme I, Breuhahn K, Moldenhauer G, Witzens-Harig M, Cerwenka A (2013) Downregulation of the activating NKp30 ligand B7-H6 by HDAC inhibitors impairs tumor cell recognition by NK cells. Blood 122 (5): 684–693.
Grillari J, Hackl M, Grillari-Voglauer R (2010) miR-17-92 cluster: ups and downs in cancer and aging. Biogerontology 11 (4): 501–506.
Gupta M, Han JJ, Stenson M, Wellik L, Witzig TE (2012) Regulation of STAT3 by histone deacetylase-3 in diffuse large B-cell lymphoma: implications for therapy. Leukemia 26 (6): 1356–1364.
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435 (7043): 828–833.
Hou ZH, Han QJ, Zhang C, Tian ZG, Zhang J (2014) miR146a impairs the IFN-induced anti-HBV immune response by downregulating STAT1 in hepatocytes. Liver Int 34 (1): 58–68.
Ji M, Rao E, Ramachandrareddy H, Shen Y, Jiang C, Chen J, Hu Y, Rizzino A, Chan WC, Fu K, McKeithan TW (2011) The miR-17-92 microRNA cluster is regulated by multiple mechanisms in B-cell malignancies. Am J Pathol 179 (4): 1645–1656.
Khan SN, Khan AU (2010) Role of histone acetylation in cell physiology and diseases: an update. Clin Chim Acta 411 (19-20): 1401–1411.
Kim MS, Kwon HJ, Lee YM, Baek JH, Jang JE, Lee SW, Moon EJ, Kim HS, Lee SK, Chung HY, Kim CW, Kim KW (2001) Histone deacetylases induce angiogenesis by negative regulation of tumor suppressor genes. Nat Med 7 (4): 437–443.
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23 (20): 4051–4060.
Li BQ, Yu H, Wang Z, Ding GH, Liu L (2013) MicroRNA mediated network and DNA methylation in colorectal cancer. Protein Pept Lett 20 (3): 352–363.
Luesink M, Hollink IH, van der Velden VH, Knops RH, Boezeman JB, de Haas V, Trka J, Baruchel A, Reinhardt D, van der Reijden BA, van den Heuvel-Eibrink MM, Zwaan CM, Jansen JH (2012) High GATA2 expression is a poor prognostic marker in pediatric acute myeloid leukemia. Blood 120 (10): 2064–2075.
Mateen S, Raina K, Jain AK, Agarwal C, Chan D, Agarwal R (2012) Epigenetic modifications and p21-cyclin B1 nexus in anticancer effect of histone deacetylase inhibitors in combination with silibinin on non-small cell lung cancer cells. Epigenetics 7 (10): 1161–1172.
Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, Akiyama M, Chauhan D, Munshi N, Gu X, Bailey C, Joseph M, Libermann TA, Richon VM, Marks PA, Anderson KC (2004) Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci USA 101 (2): 540–545.
Nagel S, Venturini L, Przybylski GK, Grabarczyk P, Schmidt CA, Meyer C, Drexler HG, Macleod RA, Scherr M (2009) Activation of miR-17-92 by NK-like homeodomain proteins suppresses apoptosis via reduction of E2F1 in T-cell acute lymphoblastic leukemia. Leuk Lymphoma 50 (1): 101–108.
Nimmanapalli R, Fuino L, Bali P, Gasparetto M, Glozak M, Tao J, Moscinski L, Smith C, Wu J, Jove R, Atadja P, Bhalla K (2003) Histone deacetylase inhibitor LAQ824 both lowers expression and promotes proteasomal degradation of Bcr-Abl and induces apoptosis of imatinib mesylate-sensitive or -refractory chronic myelogenous leukemia-blast crisis cells. Cancer Res 63 (16): 5126–5135.
Noh EJ, Lee JS (2003) Functional interplay between modulation of histone deacetylase activity and its regulatory role in G2-M transition. Biochem Biophys Res Commun 310 (2): 267–273.
O'Connell RM, Rao DS, Baltimore D (2012) microRNA regulation of inflammatory responses. Annu Rev Immunol 30: 295–312.
O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435 (7043): 839–843.
Ota A, Tagawa H, Karnan S, Tsuzuki S, Karpas A, Kira S, Yoshida Y, Seto M (2004) Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 64 (9): 3087–3095.
Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A (2008) E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13 (3): 272–286.
Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, Rameh L, Loda M, Pandolfi PP (2010) Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal 3 (117): ra29.
Qu J, Hou Z, Han Q, Jiang W, Zhang C, Tian Z, Zhang J (2014) Intracellular poly(I:C) initiated gastric adenocarcinoma cell apoptosis and subsequently ameliorated NK cell functions. J Interferon Cytokine Res 34 (1): 52–59.
Richon VM, Sandhoff TW, Rifkind RA, Marks PA (2000) Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA 97 (18): 10014–10019.
Sato Y (2013) Is histone deacetylase-9-MicroRNA-17∼92 cluster a novel axis for angiogenesis regulation? Arterioscler Thromb Vasc Biol 33 (3): 445–446.
Scian MJ, Carchman EH, Mohanraj L, Stagliano KE, Anderson MA, Deb D, Crane BM, Kiyono T, Windle B, Deb SP, Deb S (2008) Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene 27 (18): 2583–2593.
Setiadi AF, Omilusik K, David MD, Seipp RP, Hartikainen J, Gopaul R, Choi KB, Jefferies WA (2008) Epigenetic enhancement of antigen processing and presentation promotes immune recognition of tumors. Cancer Res 68 (23): 9601–9607.
Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O (2008) Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol 9 (9): 1065–1073.
Struhl K (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 12 (5): 599–606.
Su C, Hou Z, Zhang C, Tian Z, Zhang J (2011) Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J 8: 354.
Sun X, Zhang J, Wang L, Tian Z (2008) Growth inhibition of human hepatocellular carcinoma cells by blocking STAT3 activation with decoy-ODN. Cancer Lett 262 (2): 201–213.
Takada S, Asahara H (2012) Current strategies for microRNA research. Mod Rheumatol 22 (5): 645–653.
Ting CN, Olson MC, Barton KP, Leiden JM (1996) Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 384 (6608): 474–478.
Woods K, Thomson JM, Hammond SM (2007) Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem 282 (4): 2130–2134.
Yan HL, Xue G, Mei Q, Wang YZ, Ding FX, Liu MF, Lu MH, Tang Y, Yu HY, Sun SH (2009) Repression of the miR-17-92 cluster by p53 has an important function in hypoxia-induced apoptosis. EMBO J 28 (18): 2719–2732.
Yang H, Xu W, Li Y, Lan P, Zhang J, Zhang Y, Zhang C (2014) Superior activity of a new histone deacetylase inhibitor (ZYJ-34c) in inhibiting growth of human leukemia cells by induction p21WAF1 expression and cell cycle arrest. Anticancer Drugs 25 (7): 767–777.
You JS, Kang JK, Lee EK, Lee JC, Lee SH, Jeon YJ, Koh DH, Ahn SH, Seo DW, Lee HY, Cho EJ, Han JW (2008) Histone deacetylase inhibitor apicidin downregulates DNA methyltransferase 1 expression and induces repressive histone modifications via recruitment of corepressor complex to promoter region in human cervix cancer cells. Oncogene 27 (10): 1376–1386.
Zhang C, Wang Y, Zhou Z, Zhang J, Tian Z (2009) Sodium butyrate upregulates expression of NKG2D ligand MICA/B in HeLa and HepG2 cell lines and increases their susceptibility to NK lysis. Cancer Immunol Immunother 58 (8): 1275–1285.
Zhang Y, Lin A, Zhang C, Tian Z, Zhang J (2014) Phosphorothioate-modified CpG oligodeoxynucleotide (CpG ODN) induces apoptosis of human hepatocellular carcinoma cells independent of TLR9. Cancer Immunol Immunother 63 (4): 357–367.
Zhao ZN, Bai JX, Zhou Q, Yan B, Qin WW, Jia LT, Meng YL, Jin BQ, Yao LB, Wang T, Yang AG (2012) TSA suppresses miR-106b-93-25 cluster expression through downregulation of MYC and inhibits proliferation and induces apoptosis in human EMC. PLoS One 7 (9): e45133.
Acknowledgements
This work was supported by grants from the National 973 Basic Research Program of China (#2013CB944901), the Natural Science Foundation of China (#81273220, #81472646, #31200651), and the Program for Changjiang Scholars and Innovative Research Team in University (#IRT13028).
Author information
Authors and Affiliations
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Yang, H., Lan, P., Hou, Z. et al. Histone deacetylase inhibitor SAHA epigenetically regulates miR-17-92 cluster and MCM7 to upregulate MICA expression in hepatoma. Br J Cancer 112, 112–121 (2015). https://doi.org/10.1038/bjc.2014.547
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2014.547
Keywords
This article is cited by
-
Dysregulation of noncoding RNA in chordoma; implications in identifying potential targets for novel therapeutic approaches
Molecular Biology Reports (2024)
-
Novel role of immune-related non-coding RNAs as potential biomarkers regulating tumour immunoresponse via MICA/NKG2D pathway
Biomarker Research (2023)
-
HDAC Inhibitor SAHA Alleviates Pyroptosis by up-regulating miR-340 to Inhibit NEK7 Signaling in Subarachnoid Hemorrhage
Neurochemical Research (2023)
-
MiR-129-5p exerts Wnt signaling-dependent tumor-suppressive functions in hepatocellular carcinoma by directly targeting hepatoma-derived growth factor HDGF
Cancer Cell International (2022)
-
Association of MICA gene polymorphisms with thionamide-induced agranulocytosis
Journal of Endocrinological Investigation (2021)