Abstract
Dynamic variations in mitochondrial shape have been related to function. However, tools to automatically classify and enumerate mitochondrial shapes are lacking, as are systematic studies exploring the relationship of such shapes to mitochondrial stress. Here we show that during increased generation of mitochondrial reactive oxygen species (mtROS), mitochondria change their shape from tubular to donut or blob forms, which can be computationally quantified. Imaging of cells treated with rotenone or antimycin, showed time and dose-dependent conversion of tubular forms to donut-shaped mitochondria followed by appearance of blob forms. Time-lapse images showed reversible transitions from tubular to donut shapes and unidirectional transitions between donut and blob shapes. Blobs were the predominant sources of mtROS and appeared to be related to mitochondrial-calcium influx. Mitochondrial shape change could be prevented by either pretreatment with antioxidants like N-acetyl cysteine or inhibition of the mitochondrial calcium uniporter. This work represents a novel approach towards relating mitochondrial shape to function, through integration of cellular markers and a novel shape classification algorithm.
Similar content being viewed by others
Main
Mitochondria are highly dynamic organelles1, 2 and attain different shapes3 reflecting different cellular states, during the lifetime of a cell.4 Shape is an important determinant of function,1, 5 which is precisely reflected by mitochondrial dynamics.6 Recent evidences have explored the importance of mitochondrial dynamics in calcium signaling, ATP synthesis, oxidative stress, cell survival and cell-death pathways.7 In a cell, even under stasis, mitochondria continually fuse, divide and move, with the help of motor and adapter proteins.8 A huge stride has been made during the last decade, in deciphering mitochondrial dynamics9 and hence highly variable and distinct shapes of mitochondria have been observed during mitochondrial fusion and fission processes.10, 11 In most mammalian cells, mitochondria are tubular in shape under normal conditions12 but may attain various forms during various cellular perturbations. During apoptosis mitochondria are extensively fragmented and form small punctuate or round structures,13 while during necrosis mitochondria usually swell and become distended.14 During autophagy, mitochondria become elongated and are spared from degradation. Besides being tubular, swollen or elongated, mitochondria have also been shown to be ‘donut’ shaped, which is a recently discovered form of mitochondria with biochemical and pathophysiological significance.15 This diversification in shape of mitochondria makes this organelle a highly versatile in its function for maintaining cell survival and tissue homeostasis. Whether this shape diversity correlates with mitochondrial reactive oxygen species (mtROS) production, an important source of oxidative stress or to the redox state of the cell remains to be clearly established.
Despite the significant achievements in deciphering the dynamics of mitochondrial shape, there are no reports of quantification of these shapes in a single cell as a predictive marker of mitochondrial stress. Manual methods of counting different mitochondrial shapes in a single cell16 are laborious, time consuming and inefficient, which in most of the cases remain dependent on the images acquired with confocal microscopy, with proper z stacking. This also limits the use of images taken with normal fluorescent microscopes, which will be difficult to use for quantification. Automatic pattern recognition tools, which can accurately distinguish between different shapes of mitochondria, can be extremely useful in this direction and will be useful in the actively growing field of mitochondrial dynamics.
Here we show that mitochondrial shape is an important determinant of mitochondrial function in terms of ROS generation. We also report that mitochondrial calcium is an important mediator of mitochondrial blob/donut shape formation and blocking mitochondrial calcium uptake prevents the formation of blob/donut shaped mitochondria. We also developed an automated tool for simple and efficient mitochondrial shape classification to quantify stress and its correlation with the levels of mtROS generated in the cell.
Results
Mitochondrial shapes are related to mitochondrial ROS generation
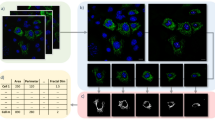
To determine the relationships between mitochondrial shapes and mtROS, mitochondria were labeled by dyes or mitochondrial specific GFP proteins (mGFP), as described in methods. Mitochondria specific ROS measuring dye (MitoSOX Red) was used to measure mtROS by flow cytometry or confocal microscopy. Mitochondrial complex I and complex III inhibitors (rotenone and antimycin), were used to induce mtROS. Dose and time-dependent increase in mtROS generation was observed in BEAS-2B cells after induction with either rotenone or antimycin (Figure 1a–d). Mitochondria were found to be mostly in tubular form in a healthy cell, but under stress conditions associated with increased mtROS, mitochondria were observed acquiring a more donut or blob form (Figure 2a). These three different shapes of mitochondria, referred to as tubular, blob or donut, were identifiable corresponding to branching tubes, circular tubes with clear center, and round respectively (Figure 2a). A time-dependent increase in donut/blob-shaped mitochondria was observed with either rotenone (Figure 2b) or antimycin treatment (Supplementary Figure S1).
Increased mitochondrial ROS generation with mitochondrial inhibitors. ROS measurement in BEAS-2B cells with different concentrations of rotenone as indicated (a). All measurements were at 12 h post rotenone. (b) Time dependent increase in ROS generation by mitochondria, by inducing cells with a sublethal rotenone concentration (100 nM). Similar Results were obtained with antimycin, which was used at different concentrations as indicated on box plots (c) and a suboptimal concentration (0.1 μM) was used for time kinetic experiments (d). MitoSOX Red fluorescence is shown on X-axis, and cell size on Y-axis (FSC)
Mitochondrial shape change with Stress. (a) Tubular mitochondrial shapes are seen in control (Con) cells, with other shapes (Donut/Blob) emerging after rotenone (Rot) treatment. A magnified portion of control or rotenone treated cell showing all the three shapes (a). (b) Time-dependent change in mitochondrial shape after rotenone treatment, with time points as indicated on figures, magnified portions of the images show a clear shape change in mitochondria with time. BEAS-2B cells were transfected with mGFP and imaged at the indicated time points. Scale bars; 10 μm
Live cell imaging was done to monitor mtROS generation and shape change in a single cell, by costaining with mitotracker green/mGFP and MitoSOX Red. BEAS-2B cells, pretreated with rotenone or antimycin were imaged under live cell conditions and further induced with 10 μM of rotenone or antimycin to measure live changes in mitochondrial shape and mtROS levels (movies 1–4). In both healthy control cells and stressed cells, tubular mitochondria were not the major source of mtROS (low fluorescence with MitoSOX red) (Figure 3a). Circular mitochondria were seen as highly fluorescent on both green (mitochondrial label) and red (mtROS) channels, seen as yellow on the overlay (Figure 3a).
Blob shaped mitochondria has increased mtROS and is an irreversible form. Blob/Donut from was associated with increased mtROS generation as revealed by live cell imaging (a). Images are shown for each color and merged for control and rotenone (100 nM for 12 h) pretreated cells at 0 s and 300 s after induction with further 10 μM of rotenone. Yellow color denotes mitochondria that are actively producing mtROS. (b) Spatial differences in mtROS quantified for different shapes of mitochondria and represented as total flourescent intensity of MitoSOX Red (FMitoSOX Red) in arbitrary units (AU). Blob shaped mitochondria were the predominant source of mtROS (b). Time lapse images of a single mitochondrion under control (c, d) and rotenone treated conditions (e), with shape change from Tubular to Blob via Donut form (e). Mitochondria are labeled with Mitotracker Green (mito Green) and mtROS (MitoSOX Red, red) (a) and with mitotracker red (c–e). Data is the representative of three different experiments with at least 10 images used for quantitation. Scale bars; 10 μm (a) and 5 μm (c–e). *denotes P<0.05 versus Tubular form
This was not restricted to BEAS-2B cells only, but similar findings were observed with alveolar epithelial cells (A549), fibrosarcoma cells (HT1080) and human primary bronchial epithelial cells (Supplementary Figure S2). Interestingly, quantitation of mtROS in all three forms of mitochondria revealed very low levels of mtROS in tubular shape, mild in the donut shape and increased mtROS in the blob shaped mitochondria (Figure 3b). Time-lapse experiments showed reversible transitions from tubular to donut shapes (Figures 3c and d), under control conditions and unidirectional transitions between donut and blob shapes (Figure 3e) with rotenone treatment.
Mitochondrial calcium influx and ROS are early mediators of mitochondrial shape change
In order to determine whether changes in mitochondrial shape can also be prevented by ROS scavenging agents, the antioxidant N-acetyl cysteine (NAC) was used.17 NAC pretreatment was associated with significant decrease in mtROS levels and donut/blob formation (Figure 4a and b), with more tubular form observed, compared with rotenone-induced cells that did not receive NAC. To determine whether NAC can reverse the donut/blob shaped mitochondria back to the tubular form, NAC treatment was given post rotenone (after 12 h), but no decrease in either mtROS or mitochondrial donut/blob form was observed (Figure 4c and d). These results suggested that increased ROS led to secondary irreversible events associated with changes in mitochondrial shape. To further study the cause of the mitochondrial shape change during stress, calcium, which is an initiator of mitochondria mediated cellular apoptosis18 was measured after rotenone treatment. Cytoplasmic calcium levels were measured in BEAS-2B cells by confocal imaging using Fluo-4AM calcium-sensitive dye. Time-dependent increase in calcium levels were observed in rotenone treated BEAS-2B cells, and by 24 h post-treatment, substantial increases in cytoplasmic calcium levels were observed (Figure 5a and b), which was also confirmed by flow cytometry (Figure 5c). Mitochondria targeted calcium GFP (inverse pericam, i-pericam), which shows diminished fluorescence with increased calcium,19 was used to study changes in mitochondrial calcium. Calcium fluorescence measured by either flow cytometry or confocal imaging, revealed increase in mitochondrial calcium by rotenone treatment in a time-dependent manner (Figure 5d–f), which corresponded to changes in mitochondrial shapes. To determine whether prevention of mitochondrial calcium uptake could prevent the blob/donut form, ruthenium360 (Ru360) was used to inhibit the mitochondrial calcium uniporter. Interestingly, it was observed that pretreatment of BEAS-2B cells with Ru360 (30 min before induction with 10 μM of rotenone) prevented mitochondrial calcium uptake and donut/blob formation, indicating mitochondrial calcium uptake as one of the important mediators of mitochondrial shape change (Figure 5g and Supplementary Figure S3). The effect was temporary and further imaging at 24 h post induction of rotenone did not show any protection (Supplementary Figure S4), suggesting that mechanisms other than uniporter mediated calcium influx are likely to be important.
Mitochondrial shape change is reversible and mediated by increased mitochondrial calcium uptake. Pretreatment of rotenone induced BEAS-2B cells with NAC prevented the formation of donut or blob-shaped mitochondria with more pronounced effect on blob shape (a). Cells were transfected with mGFP and imaged after 24 h of time under control (Con), rotenone treated (Rot) and Rotenone treated with NAC pretreatment conditions (Rot+NAC 24 h). NAC treatment was given 1 h before rotenone induction. NAC treatment was also associated with decreased mtROS production (b). (c) NAC treatment after 12 h of rotenone induction did not prevent the formation of donut/blob shaped mitochondria. Cells were labeled with mitotracker red (mito Red) to see the mitochondrial shape changes. (d) ROS measurement did not show any decrease in mtROS levels with NAC treatment after 12 h of rotenone treatment as revealed by FACS data for the mtROS. Scale bars; 10 μm
Calcium-dependent changes in mitochondrial shape. Calcium was measured in rotenone induced BEAS-2B cells with calcium sensitive dye (Fluo-4AM) at various time points, with mean cellular intensity, as quantified from the confocal images (a) and fluorescence quantitation using flow cytometry (b). Mitochondrial calcium imaging as done by i-pericam (green), in control and rotenone induced conditions (c), with mean GFP calcium intensity (d). (e) FACS based measurement of mean calcium intensity at different time points with rotenone treatment. (f) Ruthenium-360 (Ru360), a mitochondrial calcium uniporter inhibitor, inhibits mitochondrial calcium entry, measured by FACS over 300 s after 10 μM rotenone. Scale bars; 25 μm. *denotes P<0.05 versus 0 hour (h) time point
Mitochondrial shape quantification in single cell by a random forest based classifier
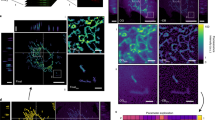
As evident from the results, donut and blob shapes were likely to be sequential stages in mitochondrial stress (Figure 6a). However, this was from measurements in relatively few cells and difficult to prove quantitatively over a large number of events. Visual scanning and quantitative assessment of large image sets produced during live confocal microscopy would require too many man-hours. Therefore, we tried to simplify quantification of mitochondrial shapes (Figure 6b) in the cells by developing a computational pipeline for recognizing and quantifying the shape changes after supervised training for shape recognition (see methods). We were able to analyze large datasets in an automated fashion and accurately distinguish between different shapes of mitochondria in a single cell through this framework (Supplementary Figure S5). Quantitation was done in a single run for more than 100 cells, under control or rotenone treated conditions at different time and the percentage of pixel count for each shape was plotted as a function of time. Figure 7c shows the original images along with their corresponding processed images, with more tubular form at lower time points and more donut or blob shape at higher time points. Consistent with the mtROS data, obtained from FACS, donut form started increasing as early as 1 h and peaked at 6 h, but there was a consistent increase in blob form, with more dramatic increase at 12 and 24 h, which contributed to overall increase in the percent total (donut+blob) form of mitochondria (Figure 7a and b). Images taken at higher magnification, in a single cell, were also trained with the software. The recognition of all the three forms of mitochondria even at higher magnification, with a slight compromise in resolution, again demonstrates the efficiency of the software-based approach in recognition of various mitochondrial shapes (Figure 7c).
Shape recognition pipeline for automated classification of different mitochondrial shapes. Graphical representation of mitochondrial shape change during stresses (a). A schematic overview of the pipeline (b). Recognition of different shapes of mitochondria by the software (c) at different time points as indicated on figures. Mitochondria were labeled with mGFP (true color) and the software processed images were pseudo colored as, blue=Tubular, white=Donut, red=Blob and green=not classifiable. Mitochondria were in Tubular form under normal conditions (cI,) with a time-dependent trend towards blob or donut form under stress conditions (cII to cVI). More than 15 images were used for quantitation. Scale bars; 10 μm
High-throughput automated mitochondrial shape quantification. Mitochondrial shape quantification done by the software shows the correlation of different shapes of mitochondria with time after rotenone treatment. (a) Percentage of mitochondria classified as belong to a particular shape, as a function of time after rotenone treatment. Tubular forms diminish with time, with donut shapes rising within 1 h and blobs increasing at late time (6 h onwards). Total refers to the sum of donut and blobs. The increase in blob mitochondrial shapes (a) inversely match the fraction of un-stressed cells (cells with low mtROS levels determined by FACS) (b). (c) High resolution confocal images to clearly visualize all three forms of mitochondria. Software could reliably distinguish between all the three forms of mitochondria, blue=Tubular, white=Donut, red=Blob and green=not detected. At least 15 images were used for quantitation. Scale bars; 5 μm
Discussion
Our study provides novel correlations of mitochondrial shape change with the level of mtROS being generated in human lung epithelial cells, and also shows how computational assessment of mitochondrial shape profiles within single cell, can serve as a marker of cell health. To the best of our knowledge, this is the first report of using supervised shape recognition algorithms in the field of mitochondrial dynamics, and can be easily adapted to other purposes. This is important because the dynamics of mitochondrial shape are now well documented,20 with the growth in understanding of the protein complexes and signaling pathways involved.21 Most of the cellular ROS is produced by mtROS, due to leakage of electrons at complexes I and III of the electron transport chain, which may lead to disruption of mitochondrial functions. We have used the mitochondrial complex I (rotenone) or complex III (antimycin) inhibitors to elevate mtROS generation in human lung epithelial cells (BEAS-2B). In order to minimize the contribution of other cytosolic ROS producing enzymes, we used mitochondria-specific ROS measuring dye, MitoSOX Red, to see the effects of mtROS on mitochondrial shape change. Sublethal concentrations of either rotenone or antimycin were selected for the study, to monitor time-dependent changes in mitochondrial shape, without leading to any significant cellular or nuclear morphological changes (Figure 2).
We observed three distinct shapes of mitochondria: Tubular, donut or blob. Time kinetic experiments showed that mitochondria are tubular in normal conditions, change to donut form under mild stress and to blob form during high oxidative stress. Notably, even an unstressed cell shows a few spontaneously forming donuts and time lapse microscopy revealed that these are in equilibrium with the tubular form (Figure 3c–e). In order to reliably quantify the changes in mitochondrial shape in a high-throughput and automated fashion, we trained a random forest based image-classification software on preprocessed images and the output was quantitated using in-house developed codes. Although there are reports of quantifying different mitochondrial shapes by manual methods,16 our computational approach is the most simple and reliable technique developed so far, which efficiently distinguished and quantified different mitochondrial shapes in a single cell or even in a portion of a cell, where the images were digitally magnified. The quantitative results confirmed the visual observations done on a smaller scale, but with more confidence, precision and accuracy (Figure 6). This approach of combining experimental and computational approaches is the first demonstration of structure-function relationship in mitochondria. Notably, we did not require the use of confocal imaging and the results were obtained from normal fluorescent microscopes without z stacking. The software was efficient in resolving the low intensity and very high intensity areas of mitochondrial images acquired without z stacking, thus making it a technique of widespread applicability (Supplementary Figure S6). Nevertheless, confocal images can be quantified in the same manner presumably yielding better results than fluorescent microscopy images. This proof of principle for structure-function relationship in mitochondria has important future implications in understanding the mitochondrial dynamics and physiology.
The relevance of these findings to physiological mechanisms of cell stress was confirmed in separate experiments where TNF-α induction was given to cells. TNF-α induction initially led to a mild increase in mtROS generation and increased donut-shaped mitochondria (Supplementary Figure S7). Donut shapes and increased mtROS were reversible if the cells were allowed to recover. This reversibility was not seen after longer TNF-α exposures, by which time the blob form with increased mtROS levels and increased apoptosis were seen (data not shown). Thus, we speculate that the donut shape of mitochondria can be an early marker of cellular stress, while the blob shape can be an early marker of irreversible toxicity. Our work is the first demonstration of using computational method to precisely quantify the degree of stress in a cell in terms of mitochondrial shape change, by using an automated tool for mitochondrial shape classification. We have also shown the relation of mitochondrial shape change to physiological parameters particularly mtROS and mitochondrial calcium, during different cellular perturbations, which has a great potential for translation.
Materials and Methods
Cell lines
BEAS-2B and human primary bronchial epithelial cells were grown in BEGM media along with the supplementary cocktail provided; A549 cells were grown in RPMI media with 10% fetal bovine serum and HT1080 were grown in DMEM low glucose with 10% fetal bovine serum. The cells were maintained under normal culture conditions with 5% CO2 and 37 °C of temperature.
Mitochondrial dyes and overexpression vectors
Mitochondrial GFP or RFP overexpression vectors (mitoGFP or mitoRFP), contain the Leader sequence of E1 alpha pyruvate dehydrogenase, with baculovirus mode of transfection; for further details, please refer to the BacMam 2.0, on LifeTechnologies website (www.invitrogen.com). The Excitation (Ex) and emission (Em) spectra of mitochondrial dyes and GFP proteins are as: mitotracker green: Ex=490 nm, Em=516 nm, mitotracker red, Ex=579 nm, Em=599 nm, mitoGFP: Ex=485 nm, Em=520 nm; mitoRFP: Ex=555 nm Em=584 nm.
Drug treatments
Rotenone, which is a mitochondrial electron transport chain complex I inhibitor and antimycin, complex III inhibitor, were purchased from sigma (Sigma Aldrich, St Louis, MO, USA). These inhibitors were used to generate mitochondrial ROS, with four different concentrations, as 10, 100, 500 or 1000 nm of rotenone, and 0.1, 1, 10, 20 μm of antimycin. Similarly TNF-α (R&D Systems, Minneapolis, MN, USA) was also used for ROS induction, at a concentration of 20 ng/ml.22 NAC, which is a ROS scavenger (Cayman, Ann Arbor, MI, USA), was used at a concentration of 100 μM.23
Immunoflourescence
Immunocytochemistry was done as described earlier.24 Briefly for mitoGFP or RFP, cells were imaged either under live conditions or fixed with paraformaldehyde and washed with phosphate-buffered saline and mounted with DAPI.
Live cell imaging
For live cell imaging cells were maintained as per the culture conditions and stained with either mitochondria specific dyes or with mitochondrial targeted GFP expression proteins. Live cell imaging was done by either fluorescent based live cell imaging system (Leica DMI6000) or confocal microscopy (Leica SP5; Ernst-Leitz-Straße, Wetzlar, Germany), with appropriate, filters and lasers.
Flow cytometry
Flow cytometry was done by using either FACS caliber (BD Biosciences, San Jose, CA, USA) as described earlier.24 For mtROS measurement, cells were stained with MitoSOX Red for 15–20 min and trypnized, washed with phosphate-buffered saline and taken for the measurement by FACS; minimum 10 thousand cells were acquired for each sample. Fluorescence of MitoSOX Red was acquired with FL2 filter settings in FACS Caliber. For mitotracker green, cells were incubated with the dye for 15–20 minutes, trypsinized and washed with phosphate-buffered saline and then taken for FACS measurement. Similarly for mitoGFP, cells were transfected with mitoGFP for 24 h and then trypsinized, washed and detected on FACS, in both the cases FL1 settings were used in the FACS caliber.
Calcium measurements
Calcium was measured in the cells by using a calcium dye, FLUO-4AM (Life Technologies, Carlsbad, CA, USA), as per the recommended protocol. Cells were seeded as described earlier and treated with proper inducers and inhibitors and during calcium measurement, cells were trypsinized and calcium dye was used along with the calcium buffer (140 mM NaCl, 5 mM KCl, 10 mM glucose, 10 mM HEPES, 2 mM CaCl2 and 1 mM MgCl2).25 Calcium fluorescence was measured in the calcium buffer, by either confocal microscopy with quantification of at least 10 images or flow cytometry, using appropriate laser and filter settings. Mitochondrial calcium was measured by either confocal microscopy or flow cytometry using mitochondrial specific targeted calcium GFP, inverse pericam (i-pericam). This mitochondrial targeted i-pericam specifically measures mitochondrial calcium, where GFP fluorescence is inversely proportional to the mitochondrial calcium.19 For live cell experiments cells were transfected with i-pericam using Lipofectamine LTX (Life Technologies, USA), as per the protocol. After 24 h of transfection cells were trypsinised and calcium fluorescence was recorded for 5 min with each image taken at 10 s interval with or without rotenone, using confocal microscopy.
Supervised classification
We used GemIdent,26 which is a freely available Random Forests based classification tool developed in Java. GemIdent is usually used as a color based classification schema but it was also able to classify artificially constructed simple and complex shapes of the same color satisfactorily. We then added a variety of spatial noise to these shapes to mimic the biological scenario and found it to be a robust image classification system (Supplementary Figure S5). The procedure was then extended to the preprocessed images containing labeled mitochondria. We found that the area under a phenotype (described later) was more robust to misclassification errors than counts of objects of a particular phenotype. Therefore, the phenotypes of interest were trained by defining boundaries rather than object centroids. Although training and retraining the classifier for feature identification, we found that the selection of the mask radius was absolutely critical to building a good classifier. There is a trade-off between the robustness of classification and computational time depending on the number of trees specified to build the forest. The classifier was then run on the test set for classification. The software generates separate black and white image files for each phenotype (classified pixels colored as white) and an additional file with a color overlay of all the phenotypes identified in the input image (Supplementary Figure S5).
Quantitation and statistical analysis for each phenotype represented by the black and white output files from GemIdent, quantification of the white area was done through Matlab (2007a, MathWorks; Natick, MA, USA) using the command ‘bwareopen’. For each image the total area represented by each of the three phenotypes was then added to give a representation of the total mitochondrial content of the cell. The fractions of each of these phenotypes with respect to the total mitochondrial content were then subject to statistical data analysis in the R statistical programming language (R Development Core Team, 2010).
Statistical analysis
Data is shown as mean±S.E.M. and is a representative of three experiments. Statistical significance was set at P<0.05.
Abbreviations
- AU:
-
Arbitrary Unit
- FACS:
-
Fluorescence-activated cell sorting
- GFP:
-
Green fluorescent protein
- NAC:
-
N-acetylcysteine
- RFP:
-
Red fluorescent protein
- ROS:
-
Reactive oxygen species
- Rot:
-
Rotenone
- TNF-α:
-
Tumor necrosis factor alpha
References
Youle RJ, van der Bliek AM . Mitochondrial fission, fusion, and stress. Science 2012; 337: 1062–1065.
Chan DC . Mitochondria: dynamic organelles in disease, aging, and development. Cell 2006; 125: 1241–1252.
Shaw JM, Nunnari J . Mitochondrial dynamics and division in budding yeast. Trends Cell Biol 2002; 12: 178–184.
Gomes LC, Di BG, Scorrano L . During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 2011; 13: 589–598.
Shutt TE, McBride HM . Staying cool in difficult times: Mitochondrial dynamics, quality control and the stress response. Biochim Biophys Acta 2012.
Jheng HF, Tsai PJ, Guo SM, Kuo LH, Chang CS, Su IJ et al. Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol Cell Biol 2012; 32: 309–319.
Perier C, Vila M . Mitochondrial biology and Parkinson's disease. Cold Spring Harb Perspect Med 2012; 2: a009332.
Fehrenbacher KL, Yang HC, Gay AC, Huckaba TM, Pon LA . Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr Biol 2004; 14: 1996–2004.
Wells RC, Picton LK, Williams SC, Tan FJ, Hill RB . Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm. J Biol Chem 2007; 282: 33769–33775.
Chen H, Chan DC . Mitochondrial dynamics—fusion, fission, movement, and mitophagy—in neurodegenerative diseases. Hum Mol Genet 2009; 18: R169–R176.
Huang P, Galloway CA, Yoon Y . Control of mitochondrial morphology through differential interactions of mitochondrial fusion and fission proteins. PLoS One 2011; 6: e20655.
Wasilewski M, Scorrano L . The changing shape of mitochondrial apoptosis. Trends Endocrinol Metab 2009; 20: 287–294.
Arnoult D . Mitochondrial fragmentation in apoptosis. Trends Cell Biol 2007; 17: 6–12.
Laporte C, Kosta A, Klein G, Aubry L, Lam D, Tresse E et al. A necrotic cell death model in a protist. Cell Death Differ 2007; 14: 266–274.
Liu X, Hajnoczky G . Altered fusion dynamics underlie unique morphological changes in mitochondria during hypoxia-reoxygenation stress. Cell Death Differ 2011; 18: 1561–1572.
Lo SC, Hannink M . PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res 2008; 314: 1789–1803.
Balkova P, Hlavackova M, Milerova M, Neckář J, Kolář F, Novák F et al. N-acetylcysteine treatment prevents the up-regulation of MnSOD in chronically hypoxic rat hearts. Physiol Res 2011; 60: 467–474.
Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R . Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell 2004; 16: 59–68.
Nagai T, Sawano A, Park ES, Miyawaki A . Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc Natl Acad Sci USA 2001; 98: 3197–3202.
Bereiter-Hahn J, Voth M . Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech 1994; 27: 198–219.
Liesa M, Palacin M, Zorzano A . Mitochondrial dynamics in mammalian health and disease. Physiol Rev 2009; 89: 799–845.
Kim JJ, Lee SB, Park JK, Yoo YD . TNF-alpha-induced ROSproduction triggering apoptosis is directly linked to Romo1 and Bcl-X(L). Cell Death Differ 2010; 17: 1420–1434.
Yedjou CG, Rogers C, Brown E, Tchounwou PB . Differential effect of ascorbic acid and n-acetyl-L-cysteine on arsenic trioxide-mediated oxidative stress in human leukemia (HL-60) cells. J Biochem Mol Toxicol 2008; 22: 85–92.
Ahmad T, Mabalirajan U, Sharma A, Aich J, Makhija L, Ghosh B et al. Simvastatin improves epithelial dysfunction and airway hyperresponsiveness: from asymmetric dimethyl-arginine to asthma. Am J Respir Cell Mol Biol 2011; 44: 531–539.
Moilanen LJ, Laavola M, Kukkonen M, Korhonen R, Leppänen T, Högestätt ED et al. TRPA1 contributes to the acute inflammatory response and mediates carrageenan-induced paw edema in the mouse. Sci Rep 2012; 2: 380 Epub.
Holmes S, Kapelner A, Lee PP . An Interactive java statistical image segmentation system. GemIdent. J Stat Softw 2009; 30: i10.
Acknowledgements
We thank Mrs Jyotirmoi Aich, Ms Suchita Singh and Mr Manish Kumar for their kind help. Mitochondrial calcium GFP (Inverse pericam plasmid) is a kind gift of Dr Mike Forte, Vollum Institute, Oregon, USA with the permission from Dr. Atsushi Miyawaki, RIKEN Brain Science Institute, Tokyo, Japan. The Research was supported by MLP 5502 from the Council of Scientific and Industrial Research, Govt. of India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Finazzi-Agro'
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Ahmad, T., Aggarwal, K., Pattnaik, B. et al. Computational classification of mitochondrial shapes reflects stress and redox state. Cell Death Dis 4, e461 (2013). https://doi.org/10.1038/cddis.2012.213
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2012.213
Keywords
This article is cited by
-
Obesity impairs cardiolipin-dependent mitophagy and therapeutic intercellular mitochondrial transfer ability of mesenchymal stem cells
Cell Death & Disease (2023)
-
Characterisation of cold-induced mitochondrial fission in porcine aortic endothelial cells
Molecular Medicine (2022)
-
Dynamic full-field optical coherence tomography allows live imaging of retinal pigment epithelium stress model
Communications Biology (2022)
-
Dynamics of mitochondrial distribution during development and asymmetric division of rice zygotes
Plant Reproduction (2022)
-
Mito Hacker: a set of tools to enable high-throughput analysis of mitochondrial network morphology
Scientific Reports (2020)