Abstract
MicroRNAs (miRNAs) have a role in the cellular defense mechanism. Nuclear factor erythroid-2-related factor 2 (Nrf2) increases antioxidant enzyme capacity. However, miRNA transcriptionally controlled by Nrf2 had been uncharacterized. Here we report that miR-125b is transactivated by Nrf2 and inhibits aryl hydrocarbon receptor (AhR) repressor (AhRR). Bioinformatic approaches enabled us to extract six candidate miRNAs. Of them, only miR-125b was increased in the kidney of mice treated with oltipraz. Nrf2 overexpression enhanced primary, precursor and mature miR-125b levels. Functional assays revealed MIR125B1 is a bona fide target gene of Nrf2. Oltipraz treatment protected the kidney from cisplatin toxicity with increase of miR-125b. Consistently, Nrf2 knockout abrogated an adaptive increase of miR-125b elicited by cisplatin, augmenting kidney injury. An integrative network of miRNA and messenger RNA changes enabled us to predict miR-125b as an inhibitor of AhRR for the control of AhR activity and cell survival. In our molecular study, miR-125b inhibited AhRR and thereby activated AhR, leading to the induction of mdm2. Consistently, p53 activation by cisplatin was diminished by either miR-125b or oltipraz treatment. The results of experiments using miR-125b mimic or small interfering RNA of AhRR verified the role of miR-125b in AhRR regulation for kidney protection. In conclusion, miR-125b is transcriptionally activated by Nrf2 and serves as an inhibitor of AhRR, which contributes to protecting kidney from acute injury.
Similar content being viewed by others
Main
MicroRNAs (miRNAs) have a role in diverse pathological states, including oxidative stress, tissue injury, and metabolic disorder.1, 2, 3 Notably, change in miRNA levels may be of help to increase the adaptive capacity of cells to pathologic insults.4, 5 Primary miRNAs (pri-miRNAs) are transcribed by RNA polymerase II-like messenger RNA (mRNA) transcription and are subsequently processed to miRNAs by drosha and dicer.6 Transcriptional induction of pri-miRNA is the crucial step for the control of total miRNA content. Transcription factors such as p53, NF-κB, and C/EBPβ participate in the transcription of miR-34a, miR-155, and miR-143.7, 8, 9 It has been shown that oxidative stress induced by toxicant changed a profiling of miRNA expression levels.10 However, the specific miRNA induced by oxidative stress and the responsible transcription factor(s) were uncharacterized.
Nuclear factor erythroid-2-related factor 2 (Nrf2) induces the expression of antioxidant genes and phase II enzymes by binding to the antioxidant response elements (ARE) comprised within their promoter regions, contributing to cellular defense mechanisms.11, 12 The cell-surviving and/or cell cycle-progressing effect of Nrf2 is not entirely explained by the ability of Nrf2 to induce anti-oxidant genes,13, 14 because many of pro-oxidants or toxicants also have a stimulating effect on Nrf2 but inhibit cell viability by controlling the fate of cells.15, 16 In addition, no information is available on the function of Nrf2 in regulating a central mediator responsible for both the control of cell cycle and the expression of metabolizing enzymes. miRNAs have the features to control cellular redox homeostasis, viability, and proliferation.4, 5, 17, 18 It has been shown that Nrf2 alters miR-1 and miR-206 levels via modulation of ROS contents.19 However, whether Nrf2 directly regulates miRNA gene transcription was not clear.
The kidney is an organ that is particularly vulnerable to drugs or toxicants due to its anatomical, physiological, and biochemical features.20, 21 In fact, acute kidney injury is one of the major dose-limiting adverse effects of drugs such as cisplatin.22 Cisplatin has been used as a model agent because of its composite nephrotoxicity (i.e., persistent inflammation, multiple cell death pathways, and inactivation of survival pathways). In view of the lack of an understanding on miRNA controlled by oxidative stress, this study investigated the levels of miRNAs increased by Nrf2 in the kidney and elucidated the functional role of specific miRNA in the survival of renal cells against cisplatin toxicity. In addition, we used a network integrating miRNA and mRNA changes to predict a target regulated by the miRNA identified. Here we report that miR-125b is transcriptionally regulated by Nrf2 and serves as an inhibitor of aryl hydrocarbon receptor (AhR) repressor (AhRR) in conjunction with the control of AhR and p53 signaling pathway, which may fill the missing link between Nrf2 and AhR crosstalk in association with cell survival. Our findings showing the novel transcriptional role of Nrf2 in the expression of MIR125B1 gene and the function of miR-125b in protecting the kidney from cisplatin toxicity may provide key information in understanding kidney pathobiology and designing a strategy for the treatment of kidney disease.
Results
Increases in miR-125b levels by Nrf2 activation
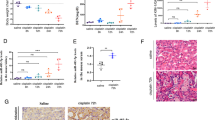
Analysis of the miRNA microarray data set (GSE19540) enabled us to extract six miRNAs upregulated in the mouse lung by oltipraz (an Nrf2 activator).23, 24 To find kidney-specific miRNA(s) affected by Nrf2, we assayed the miRNAs using quantitative real-time PCR (qRT-PCR) assays in mice orally administered with 30 mg oltipraz/kg/day for 6 consecutive days. Of them, oltipraz treatment significantly increased miR-125b levels, but not those of miR-125a, miR-26a, let-7a, let-7b, and let-7c (Figure 1a). Consistently, treatment of NRK52E cells with sulforaphane (SFN, another activator of Nrf2) increased miR-125b (Figure 1b). Similarly, overexpression of Nrf2 exerted a similar effect in HEK293A cells (Figure 1c). We also verified a significant correlation between Nrf2 and miR-125b expression in a human kidney GEO database, GSE16441 (Liu et al.25; Figure 1d). Our results indicate that activated Nrf2 may upregulate miR-125b in the kidney.
Increase in miR-125b levels in the kidney by Nrf2. (a) qRT-PCR assays of kidney miRNA levels in mice treated with oltipraz. Mice were gavaged with 30 mg oltipraz/kg/day for 6 days and were killed 1 h after the last dose. miRNAs increased by oltipraz in the lung miRNA database (GSE19540) were subjected to analyses in the kidney. Data represent the mean±S.E.M. (significantly different as compared with vehicle treatment, **P<0.01, NS, not significant; n=4 or 5 per group). (b) Increase of miR-125b by SFN treatment. miR-125b levels were assessed in NRK52E cells treated with 20 μM SFN for 12 h. (c) Increase of miR-125b by Nrf2 overexpression. miR-125b levels were assessed in HEK293A cells transfected with pcDNA3.1 or a construct encoding for Nrf2 for 48 h. (d) Correlation between miR-125b and Nrf2 mRNA levels. miR-125b and Nrf2 transcript levels were compared using human kidney miRNA and cDNA arrays database (GSE16441). For b and c, data represent the mean±S.E.M. of three independent experiments (significantly different as compared with experimental control, **P<0.01)
ARE-mediated transcriptional induction of miR-125b
To examine whether Nrf2 transcriptionally induces miR-125b, we determined the effect of Nrf2 overexpression on the levels of primary or precursor forms of miR-125b-1 and 2. MIR125B1 and MIR125B2 genes produce the same mature miR-125b via different primary transcripts.26 qRT-PCR analyses showed that enforced expression of Nrf2 notably induced primary and precursor forms of miR-125b-1 or miR-125b-2 (Figure 2a). In the in silico promoter analysis using rVISTA, there existed a conserved putative ARE sequence in the promoter regions of MIR125B1 or MIR125B2 (Figure 2b). Consistently, Nrf2 had the ability to bind to the ARE sites located within the promoter regions, as assessed by chromatin immunoprecipitation (ChIP) assays (Figure 2c; i.e., −1242 to approximately −1233 bp and −2885 to approximately −2876 bp in MIR125B1 and MIR125B2, respectively). We also determined whether specific disruption of the ARE abolished the ability of Nrf2 to induce luciferase from the miR-125b-1 promoter reporter. Nrf2 overexpression induced luciferase from the construct (Figure 2d). However, mutant (Mut) miR-125b reporter, in which the ARE sequence was mutated, exhibited a decreased response. Our data indicates that the ARE located between −1242 and −1233 bp in the MIR125B1 gene is functionally active. These results support the idea that Nrf2 transactivates the MIR125B gene through ARE.
Transcriptional induction of miR-125b1/2 by Nrf2. (a) The effect of Nrf2 overexpression on the levels of primary or precursor forms of miR-125b-1 or 2. miRNAs levels were determined using qRT-PCR in HEK293A cells transfected with pcDNA3.1 or Nrf2 for 24 h (primary) or 48 h (precursor). Data represent mean±S.E.M. from three or four independent experiments (significantly different as compared with experimental control, *P<0.05). (b) Analyses of putative Nrf2-binding sites in the promoter regions of MIR125B1 or 2. Evolutionary conserved Nrf2-binding sites were predicted at the upstream regions of MIR125B1 or MIR125B2 genes using rVISTA 2.0 program (0.75 position weight matrices cut-off value). (c) ChIP assays. Crosslinked protein–DNA complexes were immunoprecipitated using anti-RNA polymerase II antibody (Pol II, positive control), preimmune-IgG (negative control), or anti-Nrf2 antibody in HEK293A cells transfected with Nrf2. The DNA samples from immunoprecipitates were PCR-amplified using primers specific for the region −1466 to approximately −1198 bp of MIR125B1 (268 bp), the region −2724 to approximately −3002 bp of MIR125B2 (278 bp), and the control −118 to approximately +48 bp region (GAPDH promoter, 166 bp). One-tenth of the total input was used as a loading control. (d) pGL3-miR-125b-1 promoter luciferase assays. pGL3-miR-125b-1 reporter construct contains the promoter region comprising either WT or mutated (Mut) ARE (Nrf2-binding site). The luciferase reporter activity was measured in HEK293A cells co-transfected with Mock (pcDNA3.1) or Nrf2 for 24 h. Data represent mean±S.E.M. from three replicates (significantly different as compared with experimental control, **P<0.01)
Association of miR-125b and kidney protection against cisplatin
Next, we compared the degrees of cisplatin-induced renal injury in wild-type (WT) or Nrf2 knockout (Nrf2 KO) mice. According to our preliminary data (data not shown) and the previous report, we chose a day 3 time point after cisplatin for this experiment (at which serum creatinine level unchanged),27 because Nrf2 KO would aggravate kidney toxicity. In the histopathological assessment, a single dose of cisplatin injection elicited tubular cell death and inflammatory cell infiltration at day 3, which was aggravated by the loss of Nrf2 (Figure 3a, upper). In TUNEL (terminal transferase-mediated dUTP nick-end labeling) assays, a deficiency in Nrf2 worsened tubular cell apoptosis (Figure 3a lower). Consistently, the absence of Nrf2 augmented cisplatin increase of blood urea nitrogen (BUN) or kidney injury molecule-1 (KIM1) mRNA (a specific marker of kidney toxicity; Figure 3b, left and middle). Of note, Nrf2 KO abrogated an adaptive increase of miR-125b by cisplatin (Figure 3c).
The effect of Nrf2 KO on the increase of miR-125b in the kidney by cisplatin. (a–c) Kidney or blood samples were taken for biochemical assays at day 3 after a single injection of cisplatin to WT or Nrf2 KO mice (15 mg/kg, i.p., n=4 or 5 per group). (a) Hematoxylin and eosin (H&E) or TUNEL staining. Asterisks indicate epithelial cell cast, which represents necrosis of the tubular epithelium. TUNEL-positive cells were stained as red color (scale bar=100 μm). (b) BUN, renal KIM1 mRNA, and serum creatinine contents. (c) The effect of cisplatin treatment on miR-125b levels in the kidney. For b and c, data represent the mean±S.E.M. (significantly different as compared with WT Veh, *P<0.05, **P<0.01; or WT Cis, #P<0.05)
We then determined whether Nrf2 activation by multiple doses of oltipraz administration rescued the kidney from cisplatin toxicity at day 5 (at which serum creatinine level sufficiently increased).27 In histopathological examinations and molecular marker measurements, oltipraz treatment protected the kidney against cisplatin challenge (Figure 4a). Similarly, increases in BUN, KIM1 mRNA, and serum creatinine levels were all diminished by oltipraz treatment (Figure 4b). Moreover, cisplatin and/or oltipraz treatment increased miR-125b contents in the kidney (Figure 4c). All of these results support the notion that Nrf2 activation by overexpression or chemical means protects the kidney from cisplatin toxicity in association with the increase of miR-125b.
Association between increase of miR-125b and kidney protection by oltipraz. (a–c) Animals were treated with oltipraz (30 mg/kg) as described in the legend to Figure 1a. Cisplatin (15 mg/kg) was i.p. injected 1 h after the first dose of oltipraz (n=4–10 per group). (a) Hematoxylin and eosin (H&E) or TUNEL staining. Asterisks indicate desquamation of the tubular epithelium. TUNEL-positive cells were shown as red color (scale bar=100 μm). (b) BUN, renal KIM1 mRNA, and serum creatinine contents. (c) The effects of oltipraz and/or cisplatin treatment on miR-125b in the kidney. The levels of miR-125b in vehicle and oltipraz treatment in Figure 1a were repeatedly shown here, because all of the assays were done simultaneously. For b and c, data represent the mean±S.E.M. (significantly different as compared with Veh, *P<0.05, **P<0.01; or Cis, #P<0.05, ##P<0.01)
An integrative network of miR-125b target and clustered gene products
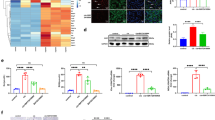
As an effort to find the target of miR-125b, complementary DNA (cDNA) microarray assays were done using the kidney tissues of WT and Nrf2 KO mice exposed to a single dose of cisplatin (intraperitoneally (i.p.), day 3 after 15 mg/kg of cisplatin treatment). In the experimental set, WT vehicle (WT-Veh) and WT cisplatin (WT-Cis) groups were included in our previous GEO database (GSE35257), because the two groups were simultaneously shared with Nrf2 KO-Veh and Nrf2 KO cisplatin (Nrf2 KO-Cis) groups in the current study (GSE48879). The genes significantly upregulated by WT-Cis relative to WT-Veh were shown as a heatmap and hierarchically correlated (Figure 5a). Moreover, hierarchical clustering using R program indicated two remarkable clusters (1 and 2). Each cluster displayed differential mRNA patterns between WT-Cis and Nrf2 KO-Cis (Figure 5b); Nrf2 KO further increased transcript levels for the genes in cluster 1 in response to cisplatin, whereas it caused the opposite effect for those in cluster 2. Gene-set enrichment analysis using DAVID program defined two significant pathways: ‘metabolism of xenobiotics cytochrome P450’ and ‘p53 signaling pathway’ (Figure 5c). In protein network analysis using GENEMANIA, the gene sets in cluster 1 and 2 were well integrated by AhR and p53 (Figure 5d). Moreover, TargetScan algorithms enabled us to select AhRR as the possible target molecule controlled by miR-125b along with AhR and p53. In the subsequent study, we focused on miR-125b regulation of AhRR for the increase in AhR activity.
An Nrf2-dependent integrative network of miR-125b and mRNA changes in gene clusters. (a) Heatmap and hierarchical correlation analyses of cDNA microarray. Microarrays were done on RNA samples extracted from the kidney at day 3 after a single injection of vehicle or cisplatin (15 mg/kg) to WT or Nrf2 KO mice. Genes significantly upregulated by WT-Cis compared with WT-Veh were shown by a heatmap with hierarchical correlation. Red, upregulation; blue, downregulation. (b) Relative mRNA levels of clustered gene sets. Fold changes relative to vehicle treatment are shown. (c) Enriched pathway analysis. Signaling pathways categorized by KEGG pathways are shown. (d) An integrative network of miR-125b, AhR, and clustered gene products. Colors indicate gene cluster. Node size reflects log2 gene expression ratio in WT mice treated with cisplatin as compared with vehicle treatment
Identification of AhRR as the inhibitory target of miR-125b
The conserved paring between AhRR 3′-untranslated region (UTR) and miR-125b sequence was nearly perfect, which was depicted with a scheme showing AhRR and AhR interaction in Figure 6a. To determine the regulatory effect of miR-125b on AhRR, immunoblotting, AhRR 3′-UTR reporter, and AhR activity reporter assays were performed after modulation of miR-125b levels. Transfection of NRK52E cells with miR-125b mimic decreased the level of AhRR (Figure 6b); sufficient AhRR expression was confirmed in the cell line (Supplementary Figure S1). miR-125b mimic transfection also diminished luciferase expression from p-EZX-AhRR 3′-UTR reporter compared with scrambled control miRNA transfection (Figure 6c). Consistently, transfection with the antisense oligonucleotide (ASO) directed against miR-125b (miR-125b-ASO) promoted luciferase expression from the construct, confirming the inhibitory effect of miR-125b on AhRR mRNA translation (Figure 6d). It was expected that miR-125b affects AhR-mediated xenobiotic response element (XRE) reporter activity because AhRR represses AhR.28 miR-125b mimic transfection indeed increased the CYP1A1 XRE reporter activity (Figure 6e). miR-125b-ASO abrogated the ability of oltipraz to increase XRE reporter activity (Figure 6f). These results provide evidence that miR-125b serves as an inhibitor of AhRR mRNA translation and increases the transcriptional activity of AhR.
Inhibition of AhRR by miR-125b for the control of AhR activity. (a) A sequence of miR-125b and its binding sites within the 3′-UTR of AhRR (left), and a scheme showing the interaction between AhRR and AhR (right). (b) miR-125b inhibition of AhRR expression. NRK52E cells were transfected with mock (scrambled control) or miR-125b mimic for 30 h. (c) miR-125b inhibition of AhRR 3′-UTR reporter activity. Luciferase activity was measured in NRK52E cells transfected with Mock (scrambled control) or miR-125b mimic in combination with a reporter comprising a luciferase cDNA fused to the 3′-UTR of AhRR (pEZX-AhRR 3′-UTR). (d) Increase of AhRR 3′-UTR reporter activity by miR-125b-ASO. Luciferase activity was similarly measured in cells transfected with control ASO (Con ASO) or miR-125b-ASO (125b ASO) in combination with pEZX-AhRR 3′-UTR. (e) CYP1A1-XRE luciferase reporter assays. Luciferase assays were done on NRK52E cells co-transfected with CYP1A1-XRE luciferase reporter, and Mock (scrambled control) or miR-125b mimic. (f) The effects of miR-125b-ASO and oltipraz on XRE reporter activity. After transfection with Con ASO or miR-125b-ASO, NRK52E cells were treated with 3 or 10 μM oltipraz for 6 h. For c–f, data represent the mean±S.E.M. of three or four independent experiments (significantly different as compared with experimental control, * P<0.05; or oltipraz-treated control, #P<0.05)
miR-125b induction of mdm2 through AhR activation
Mdm2, an inhibitor of p53, is a target gene of AhR.29 To link between AhR and p53, we examined the effect of oltipraz treatment with or without miR-125b modulation on the expression of mdm2. Oltipraz increased the levels of mdm2, beginning from 3 h to at least 24 h after treatment (Figure 7a). The ability of oltipraz to induce mdm2 was antagonized by miR-125b-ASO transfection (Figure 7b), confirming the role of miR-125b in the induction of mdm2. Consistently, oltipraz treatment abolished p53 or p21 induction by cisplatin (Figure 7c). Likewise, miR-125b mimic transfection diminished cisplatin induction of p53 (Figure 7d). AhRR knockdown had the similar effect on p53 with decreased induction of mdm2 (p53 induces mdm2; Figure 7e). Our results show that AhR activity increased by miR-125b facilitates mdm2 induction, which inhibits p53 activity.
miR-125b-dependent mdm2 induction by oltipraz for p53 repression. (a) The time-course effect of oltipraz on mdm2 expression. Immunoblottings were done on the lysates of NRK52E cells treated with 10 μM oltipraz for the indicated times. (b) The effect of miR-125b-ASO on mdm2 expression. NRK52E cells were transfected with Con ASO or miR-125b-ASO, and treated with 10 μM oltipraz for 6 h. (c) The effect of oltipraz and/or cisplatin treatment on the levels of p53 and p21. Cells were treated with 10 μM oltipraz for 12 h, followed by exposure to 30 μM cisplatin for the next 12 h. (d) The effect of miR-125b transfection on p53 and mdm2. Cells were transfected with mock (scrambled control) or miR-125b mimic, and were continuously treated with 30 μM cisplatin for the next 3 h. (e) The effect of AhRR knockdown on p53 and mdm2. NRK52E cells were similarly treated as described above after si Con or si AhRR transfection
Effect of miR-125b and AhRR on tubular cell survival
Having identified AhRR as an inhibitory target of miR-125b for p53 activity regulation, we finally explored the functional effect of miR-125b or small interfering RNA (siRNA) of AhRR on cell viability. miR-125b transfection significantly attenuated cisplatin-induced cytotoxicity of NRK52E cells (Figure 8a). Knockdown of AhRR had a similar effect (Figure 8b). As expected, oltipraz treatment improved viability of cells against cisplatin with alterations in cleaved PARP level (Figures 8c and d). We also verified that miR-125b-ASO transfection significantly diminished viability of the cells treated with cisplatin plus oltipraz compared with Con-ASO transfection (Figure 8e). These results show that miR-125b increased by Nrf2 inhibits AhRR, which may be of help to protect renal cells from toxicant-induced injury.
Increase in cell survival by miR-125b against cisplatin toxicity. (a–c, and e) Cell viability was measured using MTT assays. The values of cell viability (%) represent the mean of each group compared with the mean of vehicle treatment (100%). Data represents the mean±S.E.M. of three or four replicates (significantly different as compared with experimental control, **P<0.01; or cisplatin-treated control, #P<0.05, ##P<0.01). (a) The effect of miR-125b on cisplatin-induced cell death. NRK52E cells were transfected with scrambled control or miR-125b mimic, followed by cisplatin treatment (30 μM for 24 h). (b) Increase in cell survival by AhRR knockdown. NRK52E cells were transfected with a siRNA directed against AhRR, followed by cisplatin treatment for 24 h. (c) Increase in cell survival by oltipraz. NRK52E cells were treated with 3 or 10 μM oltipraz for 12 h followed by exposure to cisplatin for 24 h. (d) Immunoblotting for a biomarker associated with apoptosis. NRK52E cells were treated as described in the legend to Figure 7c. (e) Decrease in cell survival by miR-125b-ASO. NRK52E cells were transfected with Con ASO or miR-125b-ASO, and were similarly treated with oltipraz and cisplatin as described in c
Discussion
miRNAs have a prominent function in homeostatic plasticity, such as apoptosis, cell proliferation, and differentiation, by fine-tuning the expression of several target proteins.1, 2, 3, 4, 5 miRNA levels in the cell are controlled by several steps (i.e., transcription, processing, and stabilization); increase of pri-miRNA is the plausible approach of cells to protect from pathologic insults, because transcriptional activation by RNA polymerase II would be the major step for quantitative change of miRNAs.8, 9 In the present study, oltipraz was used as a model agent, because it has a robust effect on Nrf2; M2 metabolite from oltipraz is a strong in vivo activator of Nrf2.30, 31, 32 The results shown in this study demonstrate the ability of Nrf2 to regulate MIR125B. The causal relationship between Nrf2 and miR-125b was supported not only by the results of cell-based assays using SFN or a construct encoding for Nrf2, but by analyzing the human kidney database, implying that a quantitative variation of miR-125b by Nrf2 may have an impact on cell survival and adaptation in pathological situations accompanying oxidative stress. In the analysis of GSE19540, oltipraz treatment increased six different miRNAs in the lung. Of them, oltipraz treatment increased only miR-125b in the kidney, despite abundance of the other miRNAs; this may have resulted from organ-specific variations or different assay methods.
A mature form of miR-125b is originated from two independent genes, MIR125B1 and MIR125B2.26 An important finding of the present study is identification of the genes as the transcriptional target of Nrf2. Consistently, putative ARE sites existed in both of the promoter regions. Our observation that enforced expression of Nrf2 increased primary or precursor miR-125b-1/2 levels supports the ability of Nrf2 to transcriptionally activate the genes, which was strengthened by the presence of functionally active ARE sites. Thus, it is highly likely that Nrf2 simultaneously induces both pri-miR-125b-1 and pri-miR-125b-2. This idea was verified by the result of ARE mutation assay (MIR125B1 promoter was cloned and used for the identification of active ARE site, because the transcription start site was known).33 Mutation of the ARE significantly inhibited luciferase induction; partial inhibition may have been due to other factor(s) induced by Nrf2 or cryptic ARE site, which may exist upstream of 3 kb (analysis using rVISTA 2.0 prediction algorithms showed only one ARE site within ∼3 kb). This remains to be clarified in the future. To our knowledge, this is the first finding that Nrf2 serves as a transcriptional factor of miRNA genes.
Toxic nephropathy represents a pathological situation elicited by drugs or chemicals.34 The most representative group of chemicals that cause kidney toxicity includes cancer chemotherapeutics such as cisplatin and doxorubicin.35 Cisplatin is used for the treatment of testicular and ovarian cancers, but induces adverse effects in the kidney. Kidney injury is in fact a major problem, which limits the dose of cisplatin in clinical situations.22 Our results shown here confirmed augmented kidney toxicity in Nrf2 KO mice, as indicated by greater changes in histopathology, BUN, or KIM1 mRNA levels. The findings that cisplatin treatment increased renal miR-125b levels (presumably due to adaptive response) and which was abrogated by Nrf2 KO clearly match with the result of cell-based assays. In several cancer species, miR-125b levels are downregulated by methylation of the promoter region.36 As highly methylated promoter locus of MIR125B is in close proximity to the ARE site, the mutational activation of Nrf2, an event observed in certain malignant cancers, may not accompany increase of miR-125b. In addition, due to the epigenetic difference in MIR125B promoter between normal cell and cancer, exploitation of Nrf2 activator or other miR-125b modulator is expected to be a strategy for the rescue of kidney from cisplatin toxicity.
Nrf2 is a transcription factor necessary for the protection of many organs from oxidative stress37, 38 and is activated by a large number of beneficial compounds.39 Nrf2 facilitates the induction of target genes responsible for cell survival (i.e., Bcl-2 and Bcl-xL).40, 41 When cells are exposed to toxicants (e.g., cisplatin), Nrf2 is activated as an adaptive response to escape from oxidative stress.42 However, Nrf2 activated by toxicant would limit the capacity of cells to escape from injury, because they also stimulate cell death signaling. In our results, exposure of mice to oltipraz before cisplatin treatment protected the kidney against injury by expanding the capacity of Nrf2, which parallels the increase in miR-125b levels by oltipraz alone or in combination with cisplatin. The discrepancy between oltipraz and cisplatin in activating Nrf2 and p53 pathway would explain the differential effect on kidney cell viability despite similar increases in miR-125b by oltipraz, cisplatin, or both. Hence, it is likely that increase of miR-125b before toxicant challenge would contribute to the protection of renal cells. All of these results support the conclusion that Nrf2-dependent induction of miR-125b has a role in protecting the kidney from acute injury.
In the present study, we report an integrative network of miR-125b and mRNA changes in the cDNA arrays, which enabled us to predict AhRR as an inhibitory target of miR-125b. Moreover, clustering analyses using KEGG pathways and GENEMANIA revealed the association of AhRR with the molecules linked to antioxidant enzymes, p53, and its downstream targets. Our network raised the concept that AhRR controlled by miR-125b serves as a core molecule that balances the expression levels from two separate gene clusters. Another intriguing finding of the present study is the discovery of the fact that miR-125b increased by Nrf2 promotes the transcriptional activity of AhR. Our results from the experiments using 3′-UTR reporter, miR-125b mimic, and its inhibitor clearly demonstrate that miR-125b directly inhibits AhRR mRNA translation by binding to its 3′-UTR, which consequently activates AhR for the induction of genes comprising XRE.
AhR overactivated by environmental toxicant disrupts signaling pathways in the cell and may facilitate chemical carcinogenesis.29, 43, 44 On the other hand, complete depletion of the AhR gene also causes organ damage and carcinogenesis.45, 46, 47, 48 Thus, balanced activation of AhR by endogenous ligand seems to be required for normal physiology and cell survival. As AhR is a master transcription factor of both phase 1 and 2 enzyme genes,49 a deficiency of AhR would inhibit not only cell cycle progression and cell survival, but cellular detoxifying and antioxidant capacity, which needs be counter-balanced by the compensatory increase in antioxidant capacity. Nrf2 serves as an antioxidant transcription factor.12, 39, 50 Despite the attention on crosstalk between AhR and Nrf2,51, 52 underlying mechanisms and the physiological meaning of their functional crosstalk had been ambiguous. As AhRR represses AhR activity via competing of binding with Arnt,28 AhRR expression would cause a shift of AhR activity towards depletion. Hence, it is highly likely that Nrf2 has a role in fine-balancing between AhR and AhRR by varying miR-125b level. Our finding may fill the missing link between Nrf2 and AhR crosstalk requisite for cell survival against oxidative stress. This may also explain the beneficial impact of phytochemicals on cell survival.
Our results that oltipraz treatment induced mdm2 throughout the times examined and the ability of oltipraz to induce mdm2 was diminished by miR-125b-ASO indicate that miR-125b induces mdm2, being consistent with the ability of AhR to activate mdm2.29 Thus, an increase in AhR activity negatively regulates p53 through mdm2,29 as verified by the antagonism of oltipraz treatment or miR-125b transfection on the induction of p53 by cisplatin. This idea was strengthened by the ability of siRNA of AhRR to inhibit p53. In the present study, miR-125b transfection repressed p53 by inhibiting AhRR, which may be the major pathway for p53 inhibition in the species. Human p53 mRNA is a direct target of miR-125b. However, the 3′-UTR of mouse or rat p53 mRNA has no miR-125b-binding site.53, 54 Considering the feature of miRNA targeting diverse mRNAs associated with the molecules simultaneously linked to p53, a variation of miR-125b would effectively alter p53 activity by changing different cellular pathways. In human cells, increase of miR-125b may directly repress p53.53, 54 As the miR-125b-binding site within the 3′-UTR of AhRR is conserved in mouse, rat, and human, increase of miR-125b would have a greater impact on renal cell survival in human. Overall, it is highly likely that AhR activation, as a result of AhRR inhibition, enables cells to survive against toxic stimuli across the species.
p53 is the key molecule for the progression of apoptosis.55 In particular, activation of p53 signaling pathway is the determinant of cell death in response to toxicants.7 In Nrf2 KO mice, cisplatin treatment fortified the p53 signaling pathway, confirming the role of p53 in cell-death signaling pathway. Our results clearly showed that either miR-125b mimic or AhRR knockdown rescued cells from cisplatin-induced apoptosis. This was also accomplished by oltipraz treatment. Moreover, the rescuing effect of oltipraz was antagonized by the ASO of miR-125b, supporting the notion that the modulation of miR-125b by pharmacological activator of Nrf2 is indeed capable of protecting renal cells against toxicity. Our results also provide evidence that the regulation of miRNA may constitute an important strategy for the prevention and/or treatment of acute kidney injury. As no successful means has been developed until now to overcome kidney toxicity, our findings would provide key information on potential applications of the identified molecules for the control of cell fate and viability.
Materials and Methods
Materials
Anti-p53 antibody was purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies specifically directed against Nrf2, mdm2, p21, and PARP were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-AhRR antibody (ab85666) was purchased from Abcam (Cambridge, UK). Horseradish peroxidase-conjugated goat anti-rabbit and goat anti-mouse IgGs were obtained from Zymed Laboratories (San Francisco, CA, USA). Oltipraz was provided from CJ Corporation (Seoul, Korea), as described previously.56 Anti-β-actin antibody, cisplatin, and other reagents were supplied from Sigma (St. Louis, MO, USA).
Animal treatment
Animal experiments were conducted under the guidelines of the Institutional Animal Use and Care Committee at Seoul National University. Male C57BL/6 mice (6 weeks old) were purchased from Samtako Company (Osan, Korea), and housed at 20±2 °C with 12-h light/dark cycles and a relative humidity of 50±5% (Tecniplast, Varese, Italy) under filtered, pathogen-free air, with food (Purina, Seoul, Korea) and water available ad libitum. Mice were orally treated with multiple doses of oltipraz (30 mg/kg body weight/day), as indicated in the schemes in Figures 1 and 4. After the mice were injected with a single dose of cisplatin (i.p., 15 mg/kg body weight), kidney tissues were taken for biochemical assays as depicted in the schemes. Normal saline and 30% PEG400 were used as vehicles for cisplatin and oltipraz, respectively. Nrf2 KO mice12 supplied by RIKEN BioResource Center (Tsukuba, Japan) were bred and maintained. Nrf2 KO C57/BL6 mice were backcrossed with WT C57/BL6 mice for at least 6 months, and only male mice were used for experiments. Three days after WT and Nrf2 KO mice were injected with a single dose of cisplatin, the kidney samples were taken for biochemical assays. For serum preparation, blood was collected in BD Vacutainer serum collection tubes (BD, Franklin Lakes, NJ, USA) and was allowed to clot for 1 h at room temperature. After centrifugation at 1300 × g for 15 min, the serum supernatant was transferred to a clean tube and spun at 2700 × g. Aliquots of the samples were prepared and immediately stored at −80 °C.
Cell culture
HepG2, HEK293, and NRK52E (an epithelial type of normal rat kidney cell line) cells were obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin at 37 °C in humidified atmosphere with 5% CO2.
Transient transfection and luciferase assays
All transfection experiments were conducted using FuGENE HD Reagent (Promega, Madison, WI, USA) in accordance with manufacturer’s instruction. Cells were transfected with plasmids encoding Nrf2 (1 μg for Figures 1c, 2a and c, or 0.5 μg for Figure 2d). The empty plasmid, pcDNA3.1, was used for mock transfection. To knock down AhRR, cells were transfected with its specific siRNA (ON-TARGETplus SMARTpool L-104632-02, Dharmacon Inc., Lafayette, CO, USA). To modulate miR-125b levels, cells were transfected with miR-125b mimic (sense: 5′-UCCCUGAGACCCUAACUUGUGA-3′, antisense: 5′-ACAAGUUAGGGUCUCAGGCACU-3′) or ASO (5′-TCACAAGTTAGGGTCTCAGGGA-3′, all nucleotides were 2′-O-methyl modified; 100 nM each). The scrambled control of miR-125b (sense: 5′-GCAAUUUGGCGUCCUCCACUAA-3′ antisense: 5′-AGUGGAGGACGCCAAAUUCCCU-3′) or control ASO (an unrelated control of sequence 5′-CCTTCCCTGAAGGTTCCTCCTT-3′, all nucleotides were 2′-O-methyl modified) was used for mock transfection (100 nM each). pGL3-miR-125b-1 reporter construct contains the promoter region comprising either WT or the mutated (5′-TGACXXXGC-3′→5′-GTCCXXGC-3′) ARE (Nrf2-binding site). The plasmid containing Luc-AhRR-3′-UTR (product ID: RmiT047220-MT01; Genecopoeia, Rockville, MD, USA) was specifically synthesized. pGL-CYP1A1-1195 reporter construct containing eight XREs (AhR-binding site) was transfected to measure AhR transcription. Luciferase activity assays were performed with LUC-Pair miR Luciferase assay kit (Genocopoeia) or Luciferase assay system (Promega) according to the manufacturers’ protocols.
RNA isolation and RT-PCR assays
Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA, USA) and was reverse-transcribed. qRT-PCR was performed with the Light Cycler 1.5 (Roche, Mannheim, Germany). The relative levels of human pri-miR-125b-1 (sense: 5′-CGAACAGAAATTGCCTGTCA-3′, antisense: 5′-ACCAAATTTCCAGGATGCAA-3′); human pri-miR-125b-2 (sense: 5′-GAATTCTACCGCATCAAACCA-3′, antisense: 5′-TCTGGTGGTAACTGCAGACAA-3′); and mouse KIM1 (sense: 5′-TTCTCCCAGGCACTGTGGAT-3′, antisense: 5′-TGTTGGAATGCCAGATGAAA-3′) were normalized based on the level of glyceraldehyde-3-phosphate dehydrogenase. The relative levels of human pre-miR-125b-1 (sense: 5′-TGCGCTCCTCTCAGTCCCTGAGA-3′, antisense: 5′-GACTCGCAGCTCCCAAGA-3′); human pre-miR-125b-2 (sense: 5′-ACCAGACTTTTCCTAGTCCCT-3′, antisense: 5′-AGGTCCCAAGAGCCTGACTT-3′); miR-125b (5′-TCCCTGAGACCCTAACTTGTGA-3′); miR-125a (5′-TCCCTGAGACCCTTTAACCTGTGA-3′); miR-26a (5′-TTCAAGTAATCCAGGATAGGCT-3′); let-7a (5′-TGAGGTAGTAGGTTGTATAGTT-3′); let-7b (5′-TGAGGTAGTAGGTTGTGTGGTT-3′); and let-7c (5′-TGAGGTAGTAGGTTGTATGGTT-3′) were normalized based on the level of RNU6B. Mature miRNAs were amplified using each specific miR primer and miScript universal primer (Qiagen, Hilden, Germany). After PCR amplifications, a melting curve of each amplicon was determined to verify its accuracy.
ChIP assay
ChIP assay was done using the EZ ChIP kit (Milipore, Billerica, MA, USA) according to the manufacturer’s protocol. Nrf2 bound DNA was pulled down with anti-Nrf2 antibody (H-300) in HEK293A cells transfected with Nrf2. The DNA samples from immunoprecipitates were PCR amplified using primers specific for the MIR125B1 gene (sense: 5′-TGAGCAAGGTAGATGGCTGG-3′, antisense: 5′-AGTTGTCTTGAAGGTGGGGG-3′) or MIR125B2 promoter (sense: 5′-CTGCATGCTGGGATCTGTCT-3′, antisense: 5′-TTCGCAACACAAAAGGCAAT-3′), and the control primers (Millipore).
Histopathology
The renal cortices were fixed in 10% buffered neutral formalin for 6 h. The samples were stained with hematoxylin and eosin. TUNEL assay was also done using the DeadEnd Colorimetric TUNEL System (Promega), according to the manufacturer’s instruction. Images of renal morphology were obtained by light microscopy.
Microarray analysis and bioinformatics
The integrity of total RNA extracted from the kidney was assessed using a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). The RNA integrity numbers was 10 for all samples. Five hundred nanograms of high-quality total RNA was amplified with a TotalPrep amplification kit (Ambion, Austin, TX, USA) and 750 ng of amplified cRNA was hybridized to the Illumina MouseWG-6 expression beadchip for 16 h. The hybridized array was scanned with a BeadArray Reader (Illumina, San Diego, CA, USA). The microarray data were deposited in GEO database before submission (accession number: GSE48879). The Illumina MouseWG-6 Expression BeadChip enabled us to detect expression levels for nearly 31 000 genes in the kidney day 3 after treatment of mice with a single dose of vehicle or cisplatin. Three array replicates were obtained for each condition. The probe intensities were normalized at log2 scale using quantile normalization in beadarray 1.6 using R software (Bioconductor; http://www.bioconductor.org in the public domain). Differentially expressed genes (DEGs) by cisplatin were identified independently using Student’s t-test: DEGs were selected as the genes with Student’s t-test P-values<0.05 with a fold-change of >1.5 (Figure 5a). Hierarchical clustering with Pearson’s correlation of the DEGs was analyzed using R software (Bioconductor, pvclust). Statistically enriched signaling pathways of clustered DEGs were ranked and categorized according to KEGG pathway using DAVID 6.7 software.57 Gene interaction network between the clustered DEGs was achieved by GENEMANIA software58 and visualized by Cytoscape 3.0.0 software.59 Putative target mRNAs for miR-125b were predicted using Targetscan algorithm.
Immunoblot analysis
Cell lysates were prepared according to previously published methods.56 Protein bands of interest were developed using the ECL chemiluminescence system (Amersham, Buckinghamshire, UK). Equal loading of protein was verified by immunoblotting for β-actin.
MTT assays
The MTT assay was performed according to the previously published method.60 Absorbance was detected at 540 nm using an enzyme-linked immunosorbent assay microplate reader (Tecan, Durham, NC, USA). Cell viability was calculated relative to untreated control samples, that is, viability (% of control)=100 × (absorbance of treated sample/absorbance of control sample).60
Statistical analyses
Pearson’s coefficients with associated P-values were used for correlation analysis. For animal experiments and cell-based assays, data represent the mean±S.E.M. The differences between groups were analyzed using two-tailed Student’s t-test. The criterion for statistical significance was set at P<0.05 or P<0.01.
Accession codes
Abbreviations
- AhR:
-
aryl hydrocarbon receptor
- AhRR:
-
AhR repressor
- ARE:
-
antioxidant response element
- ASO:
-
antisense oligonucleotide
- BUN:
-
blood urea nitrogen
- cDNA:
-
complementary DNA
- ChIP:
-
chromatin immunoprecipitation
- Cis:
-
cisplatin
- KIM1:
-
kidney injury molecule-1
- KO:
-
knockout
- miRNA:
-
microRNA
- mRNA:
-
messenger RNA
- Mut:
-
mutant
- Nrf2:
-
nuclear factor erythroid-2-related factor 2
- pri-miRNA:
-
primary miRNA
- qRT-PCR:
-
quantitative real-time PCR
- SFN:
-
sulforaphane
- siRNA:
-
small interfering RNA
- UTR:
-
untranslated region
- Veh:
-
vehicle
- WT:
-
wild type
- XRE:
-
xenobiotic response element
References
Chandrasekaran K, Karolina DS, Sepramaniam S, Armugam A, Wintour EM, Bertram JF et al. Role of microRNAs in kidney homeostasis and disease. Kidney Int 2012; 81: 617–627.
Lan YF, Chen HH, Lai PF, Cheng CF, Huang YT, Lee YC et al. MicroRNA-494 reduces ATF3 expression and promotes AKI. J Am Soc Nephrol 2012; 23: 2012–2023.
Rottiers V, Naar AM . MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 2012; 13: 239–250.
Hermeking H . The miR-34 family in cancer and apoptosis. Cell Death Differ 2010; 17: 193–199.
Garofalo M, Condorelli GL, Croce CM, Condorelli G . MicroRNAs as regulators of death receptors signaling. Cell Death Differ 2010; 17: 200–208.
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 2004; 23: 4051–4060.
Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 2011; 13: 317–323.
Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 2007; 179: 5082–5089.
Jiang S, Zhang LF, Zhang HW, Hu S, Lu MH, Liang S et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J 2012; 31: 1985–1998.
Wang Z, Liu Y, Han N, Chen X, Yu W, Zhang W et al. Profiles of oxidative stress-related microRNA and mRNA expression in auditory cells. Brain Res 2010; 1346: 14–25.
Ma Q . Role of nrf2 in oxidative stress and toxicity. Ann Rev Pharmacol Toxicol 2013; 53: 401–426.
Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 1997; 236: 313–322.
Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, Haraguchi N et al. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res 2009; 15: 3423–3432.
Reddy NM, Kleeberger SR, Bream JH, Fallon PG, Kensler TW, Yamamoto M et al. Genetic disruption of the Nrf2 compromises cell-cycle progression by impairing GSH-induced redox signaling. Oncogene 2008; 27: 5821–5832.
Pabla N, Dong Z . Cisplatin nephrotoxicity: mechanisms and renoprotective strategies. Kidney Int 2008; 73: 994–1007.
Jaeschke H, Gores GJ, Cederbaum AI, Hinson JA, Pessayre D, Lemasters JJ . Mechanisms of hepatotoxicity. Toxicol Sci 2002; 65: 166–176.
Hulsmans M, De Keyzer D, Holvoet P . MicroRNAs regulating oxidative stress and inflammation in relation to obesity and atherosclerosis. FASEB J 2011; 25: 2515–2527.
Muratsu-Ikeda S, Nangaku M, Ikeda Y, Tanaka T, Wada T, Inagi R . Downregulation of miR-205 modulates cell susceptibility to oxidative and endoplasmic reticulum stresses in renal tubular cells. PLoS One 2012; 7: e41462.
Singh A, Happel C, Manna SK, Acquaah-Mensah G, Carrerero J, Kumar S et al. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest 2013; 123: 2921–2934.
Perazella MA . Renal vulnerability to drug toxicity. Clin J Am Soc Nephrol 2009; 4: 1275–1283.
Ciarimboli G, Ludwig T, Lang D, Pavenstädt H, Koepsell H, Piechota HJ et al. Cisplatin nephrotoxicity is critically mediated via the human organic cation transporter 2. Am J Pathol 2005; 167: 1477–1484.
Kelland L . The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer 2007; 7: 573–584.
Izzotti A, Calin GA, Steele VE, Cartiglia C, Longobardi M, Croce CM et al. Chemoprevention of cigarette smoke-induced alterations of MicroRNA expression in rat lungs. Cancer Prev Res 2010; 3: 62–72.
Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M . MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res 2008; 18: 404–411.
Liu H, Brannon AR, Reddy AR, Alexe G, Seiler MW, Arreola A et al. Identifying mRNA targets of microRNA dysregulated in cancer: with application to clear cell Renal Cell Carcinoma. BMC Syst Biol 2010; 4: 51.
Lee YS, Kim HK, Chung S, Kim KS, Dutta A . Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem 2005; 280: 16635–16641.
Ichimura T, Hung CC, Yang SA, Stevens JL, Bonventre JV . Kidney injury molecule-1: a tissue and urinary biomarker for nephrotoxicant-induced renal injury. Am J Physiol Renal Physiol 2004; 286: F552–F563.
Evans BR, Karchner SI, Allan LL, Pollenz RS, Tanguay RL, Jenny MJ et al. Repression of aryl hydrocarbon receptor (AHR) signaling by AHR repressor: role of DNA binding and competition for AHR nuclear translocator. Mol Pharmacol 2008; 73: 387–398.
Pääjärvi G, Viluksela M, Pohjanvirta R, Stenius U, Högberg J et al. TCDD activates Mdm2 and attenuates the p53 response to DNA damaging agents. Carcinogenesis 2005; 26: 201–208.
Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P et al. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci USA 2001; 98: 3410–3415.
Kang KW, Cho IJ, Lee CH, Kim SG . Essential role of phosphatidylinositol 3-kinase-dependent CCAAT/enhancer binding protein beta activation in the induction of glutathione S-transferase by oltipraz. J Natl Cancer Inst 2003; 95: 53–66.
Ko MS, Lee SJ, Kim JW, Lim JW, Kim SG . Differential effects of the oxidized metabolites of oltipraz on the activation of CCAAT/enhancer binding protein-beta and NF-E2-related factor-2 for GSTA2 gene induction. Drug Metab Dispos 2006; 34: 1353–1360.
Zhou R, Hu G, Liu J, Gong AY, Drescher KM, Chen XM . NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PLoS Pathog 2009; 5: e1000681.
Schreiner GE . Toxic nephropathy: adverse renal effects caused by drugs and chemicals. JAMA 1965; 191: 849–850.
Vogelzang NJ . Nephrotoxicity from chemotherapy: prevention and management. Oncology 1991; 5: 97–102 105; disc 105 109-111.
Zhang Y, Yan LX, Wu QN, Du ZM, Chen J, Liao DZ et al. miR-125b is methylated and functions as a tumor suppressor by regulating the ETS1 proto-oncogene in human invasive breast cancer. Cancer Res 2011; 71: 3552–3562.
Liu M, Grigoryev DN, Crow MT, Haas M, Yamamoto M, Reddy SP et al. Transcription factor Nrf2 is protective during ischemic and nephrotoxic acute kidney injury in mice. Kidney Int 2009; 76: 277–285.
Park HM, Cho JM, Lee HR, Shim GS, Kwak MK . Renal protection by 3H-1,2-dithiole-3-thione against cisplatin through the Nrf2-antioxidant pathway. Biochem Pharmacol 2008; 76: 597–607.
Surh YJ . Cancer chemoprevention with dietary phytochemicals. Nat Rev Cancer 2003; 3: 768–780.
Niture SK, Jaiswal AK . Nrf2-induced antiapoptotic Bcl-xL protein enhances cell survival and drug resistance. Free Radic Biol Med 2013; 57: 119–131.
Niture SK, Jaiswal AK . Nrf2 protein up-regulates antiapoptotic protein Bcl-2 and prevents cellular apoptosis. J Biol Chem 2012; 287: 9873–9886.
Aleksunes LM, Goedken MJ, Rockwell CE, Thomale J, Manautou JE, Klaassen CD . Transcriptional regulation of renal cytoprotective genes by Nrf2 and its potential use as a therapeutic target to mitigate cisplatin-induced nephrotoxicity. J Pharmacol Exper Ther 2010; 335: 2–12.
Brooks RA, Gooderham NJ, Edwards RJ, Boobis AR, Winton DJ . The mutagenicity of benzo[a]pyrene in mouse small intestine. Carcinogenesis 1999; 20: 109–114.
Tian Y, Ke S, Denison MS, Rabson AB, Gallo MA . Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J Biol Chem 1999; 274: 510–515.
Fernandez-Salguero P, Pineau T, Hilbert DM, McPhail T, Lee SS, Kimura S et al. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science 1995; 268: 722–726.
Fan Y, Boivin GP, Knudsen ES, Nebert DW, Xia Y, Puga A . The aryl hydrocarbon receptor functions as a tumor suppressor of liver carcinogenesis. Cancer Res 2010; 70: 212–220.
Xue J, Nguyen DT, Habtezion A . Aryl hydrocarbon receptor regulates pancreatic IL-22 production and protects mice from acute pancreatitis. Gastroenterology 2012; 143: 1670–1680.
Baban B, Liu JY, Mozaffari MS . Aryl hydrocarbon receptor agonist, leflunomide, protects the ischemic-reperfused kidney: role of Tregs and stem cells. Am J Physiol Regul Integr Compar Physiol 2012; 303: R1136–R1146.
Kohle C, Bock KW . Coordinate regulation of Phase I and II xenobiotic metabolisms by the Ah receptor and Nrf2. Biochem Pharmacol 2007; 73: 1853–1862.
Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA 2002; 99: 11908–11913.
Miao W, Hu L, Scrivens PJ, Batist G . Transcriptional regulation of NF-E2 p45-related factor (NRF2) expression by the aryl hydrocarbon receptor-xenobiotic response element signaling pathway: direct cross-talk between phase I and II drug-metabolizing enzymes. J Biol Chem 2005; 280: 20340–20348.
Hayes JD, Dinkova-Kostova AT, McMahon M . Cross-talk between transcription factors AhR and Nrf2: lessons for cancer chemoprevention from dioxin. Toxicol Sci 2009; 111: 199–201.
Le MT, Shyh-Chang N, Khaw SL, Chin L, Teh C, Tay J et al. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet 2011; 7: e1002242.
Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev 2009; 23: 862–876.
Fridman JS, Lowe SW . Control of apoptosis by p53. Oncogene 2003; 22: 9030–9040.
Bae EJ, Yang YM, Kim JW, Kim SG et al. Identification of a novel class of dithiolethiones that prevent hepatic insulin resistance via the adenosine monophosphate-activated protein kinase-p70 ribosomal S6 kinase-1 pathway. Hepatology 2007; 46: 730–739.
Huang da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010; 38: W214–W220.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504.
Koo JH, Lee WH, Lee CG, Kim SG . Fyn inhibition by cycloalkane-fused 1,2-dithiole-3-thiones enhances antioxidant capacity and protects mitochondria from oxidative injury. Mol Pharmacol 2012; 82: 27–36.
Acknowledgements
This research was supported by a grant (10182KFDA992) from the Ministry of Food and Drug Safety in 2010–2012.
Author Contributions
The overall study was conceived and designed by MSJ, CGL, and SGK; MSJ and CGL performed the experiments; MSJ, CGL, and JHK analyzed data; MSJ and SGK wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by E Candi
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Joo, M., Lee, C., Koo, J. et al. miR-125b transcriptionally increased by Nrf2 inhibits AhR repressor, which protects kidney from cisplatin-induced injury. Cell Death Dis 4, e899 (2013). https://doi.org/10.1038/cddis.2013.427
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.427
Keywords
This article is cited by
-
Uremic toxins mediate kidney diseases: the role of aryl hydrocarbon receptor
Cellular & Molecular Biology Letters (2024)
-
Cisplatin nephrotoxicity: new insights and therapeutic implications
Nature Reviews Nephrology (2023)
-
Plk2-mediated phosphorylation and translocalization of Nrf2 activates anti-inflammation through p53/Plk2/p21cip1 signaling in acute kidney injury
Cell Biology and Toxicology (2023)
-
Role of miR-653 and miR-29c in downregulation of CYP1A2 expression in hepatocellular carcinoma
Pharmacological Reports (2022)
-
Crosstalk between noncoding RNAs and ferroptosis: new dawn for overcoming cancer progression
Cell Death & Disease (2020)