Abstract
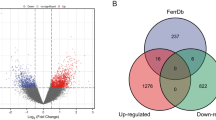
Cytogenetic analyses have revealed that complex karyotypes with numerous and highly variable genomic aberrations including single-nucleotide polymorphisms (SNPs) and copy number variants (CNVs), are observed in most of the conventional osteosarcomas (OSs). Several genome-wide studies have reported that the dysregulated expression of many genes is correlated with genomic aberrations in OS. We first compared OS gene expression in Gene Expression Omnibus (GEO) data sets and genomic aberrations in International Cancer Genome Consortium (ICGC) database to identify differentially expressed genes (DEGs) associated with SNPs or CNVs in OS. Then the function annotation of SNP- or CNV-associated DEGs was performed in terms of gene ontology analysis, pathway analysis and protein–protein interactions (PPIs). Finally, the expression of genes correlated with both SNPs and CNVs were confirmed by quantitative reverse-transcription PCR. Eight publicly available GEO data sets were obtained, and a set of 979 DEGs were identified (472 upregulated and 507 downregulated DEGs). Moreover, we obtained 1039 SNPs mapped in 938 genes, and 583 CNV sites mapped in 2915 genes. Comparing genomic aberrations and DGEs, we found 41 SNP-associated DEGs and 124 CNV-associated DEGs, in which 7 DGEs were associated with both SNPs and CNVs, including WWP1, EXT1, LDHB, C8orf59, PLEKHA5, CCT3 and VWF. The result of function annotation showed that ossification, bone development and skeletal system development were the significantly enriched terms of biological processes for DEGs. PPI network analysis showed that CCT3, COPS3 and WWP1 were the significant hub proteins. We conclude that these genes, including CCT3, COPS3 and WWP1 are candidate driver genes of importance in OS tumorigenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Kansara M, Teng MW, Smyth MJ, Thomas DM . Translational biology of osteosarcoma. Nat Rev Cancer 2014; 14: 722–735.
Moore DD, Luu HH . Osteosarcoma. Cancer Treat Res 2014; 162: 65–92.
Ta HT, Dass CR, Choong PF, Dunstan DE . Osteosarcoma treatment: state of the art. Cancer Metastasis Rev 2009; 28: 247–263.
Zhang J, Yu XH, Yan YG, Wang C, Wang WJ . PI3K/Akt signaling in osteosarcoma. Clin Chim Acta 2015; 444: 182–192.
Lin CH, Ji T, Chen CF, Hoang BH . Wnt signaling in osteosarcoma. Adv Exp Med Biol 2014; 804: 33–45.
Yan J, Wang Q, Zou K, Wang L, Schwartz EB, Fuchs JR et al. Inhibition of the JAK2/STAT3 signaling pathway exerts a therapeutic effect on osteosarcoma. Mol Med Rep 2015; 12: 498–502.
Tao J, Jiang MM, Jiang L, Salvo JS, Zeng HC, Dawson B et al. Notch activation as a driver of osteogenic sarcoma. Cancer Cell 2014; 26: 390–401.
Boehm A, Neff J, Squire J, Bayani J, Nelson M, Bridge J . Cytogenetic findings in 36 osteosarcoma specimens and a review of the literature. Fetal Pediatr Pathol 2000; 19: 359–376.
Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet 2010; 6: e1000952.
Jarvinen AK, Autio R, Haapa-Paananen S, Wolf M, Saarela M, Grenman R et al. Identification of target genes in laryngeal squamous cell carcinoma by high-resolution copy number and gene expression microarray analyses. Oncogene 2006; 25: 6997–7008.
Skotheim RI, Autio R, Lind GE, Kraggerud SM, Andrews PW, Monni O et al. Novel genomic aberrations in testicular germ cell tumors by array-CGH, and associated gene expression changes. Cell Oncol 2006; 28: 315–326.
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 2013; 41: D991–D995.
Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007; 8: R183.
Young MD, Wakefield MJ, Smyth GK, Oshlack A . Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010; 11: R14.
Mao X, Cai T, Olyarchuk JG, Wei L . Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005; 21: 3787–3793.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 2498–2504.
Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M . BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006; 34: D535–D539.
Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res 2013; 41: D816–D823.
Klevebring D, Fagerberg L, Lundberg E, Emanuelsson O, Uhlen M, Lundeberg J . Analysis of transcript and protein overlap in a human osteosarcoma cell line. BMC Genomics 2010; 11: 684.
Kuijjer ML, Hogendoorn PC, Cleton-Jansen AM . Genome-wide analyses on high-grade osteosarcoma: making sense of a genomically most unstable tumor. Int J Cancer 2013; 133: 2512–2521.
Poos K, Smida J, Maugg D, Eckstein G, Baumhoer D, Nathrath M et al. Genomic heterogeneity of osteosarcoma - shift from single candidates to functional modules. PLoS One 2015; 10: e0123082.
Sun L, Li J, Yan B . Gene expression profiling analysis of osteosarcoma cell lines. Mol Med Rep 2015; 12: 4266–4272.
Yang JL . Investigation of osteosarcoma genomics and its impact on targeted therapy: an international collaboration to conquer human osteosarcoma. Chin J Cancer 2014; 33: 575–580.
Verdecia MA, Joazeiro CA, Wells NJ, Ferrer JL, Bowman ME, Hunter T et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell 2003; 11: 249–259.
Cheng Q, Cao X, Yuan F, Li G, Tong T . Knockdown of WWP1 inhibits growth and induces apoptosis in hepatoma carcinoma cells through the activation of caspase3 and p53. Biochem Biophys Res Commun 2014; 448: 248–254.
Sarrion P, Sangorrin A, Urreizti R, Delgado A, Artuch R, Martorell L et al. Mutations in the EXT1 and EXT2 genes in Spanish patients with multiple osteochondromas. Sci Rep 2013; 3: 1346.
Philibert RA, Nelson JJ, Sandhu HK, Crowe RR, Coryell WH . Association of an exonic LDHA polymorphism with altered respiratory response in probands at high risk for panic disorder. Am J Med Genet B Neuropsychiatr Genet 2003; 117B: 11–17.
Henriksen J, Aagesen TH, Maelandsmo GM, Lothe RA, Myklebost O, Forus A . Amplification and overexpression of COPS3 in osteosarcomas potentially target TP53 for proteasome-mediated degradation. Oncogene 2003; 22: 5358–5361.
Midorikawa Y, Tsutsumi S, Taniguchi H, Ishii M, Kobune Y, Kodama T et al. Identification of genes associated with dedifferentiation of hepatocellular carcinoma with expression profiling analysis. Jpn J Cancer Res 2002; 93: 636–643.
Skawran B, Steinemann D, Weigmann A, Flemming P, Becker T, Flik J et al. Gene expression profiling in hepatocellular carcinoma: upregulation of genes in amplified chromosome regions. Mod Pathol 2008; 21: 505–516.
Both J, Wu T, Bras J, Schaap GR, Baas F, Hulsebos TJ . Identification of novel candidate oncogenes in chromosome region 17p11.2-p12 in human osteosarcoma. PLoS One 2012; 7: e30907.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Xiong, Y., Wu, S., Du, Q. et al. Integrated analysis of gene expression and genomic aberration data in osteosarcoma (OS). Cancer Gene Ther 22, 524–529 (2015). https://doi.org/10.1038/cgt.2015.48
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cgt.2015.48
This article is cited by
-
The role of WWP1 and WWP2 in bone/cartilage development and diseases
Molecular and Cellular Biochemistry (2024)
-
Dog breeds and body conformations with predisposition to osteosarcoma in the UK: a case-control study
Canine Medicine and Genetics (2021)
-
The emerging role of WWP1 in cancer development and progression
Cell Death Discovery (2021)
-
Integration of genomic copy number variations and chemotherapy-response biomarkers in pediatric sarcoma
BMC Medical Genomics (2019)
-
Elucidating the transcriptional program of feline injection-site sarcoma using a cross-species mRNA-sequencing approach
BMC Cancer (2019)