Abstract
Coronaviruses have developed various measures to evade innate immunity. We have previously shown that severe acute respiratory syndrome (SARS) coronavirus M protein suppresses type I interferon (IFN) production by impeding the formation of functional TRAF3-containing complex. In this study, we demonstrate that the IFN-antagonizing activity is specific to SARS coronavirus M protein and is mediated through its first transmembrane domain (TM1) located at the N terminus. M protein from human coronavirus HKU1 does not inhibit IFN production. Whereas N-linked glycosylation of SARS coronavirus M protein has no influence on IFN antagonism, TM1 is indispensable for the suppression of IFN production. TM1 targets SARS coronavirus M protein and heterologous proteins to the Golgi apparatus, yet Golgi localization is required but not sufficient for IFN antagonism. Mechanistically, TM1 is capable of binding with RIG-I, TRAF3, TBK1 and IKKε, and preventing the interaction of TRAF3 with its downstream effectors. Our work defines the molecular architecture of SARS coronavirus M protein required for suppression of innate antiviral response.
Similar content being viewed by others
Introduction
Host cells combat invading viruses by triggering innate immune response. As a counter-defense, viruses encode various immunosuppressive proteins to evade innate immunity. This interaction between host cells and viruses dictates the outcome of viral infection.1
Coronaviruses are enveloped and positive-stranded RNA viruses with a large genome of ∼30 kb. In the family Coronaviridae, there are four genera. In addition to human coronaviruses 229E and NL63 in the genus Alphacoronavirus, four other viruses belonging to the genus Betacoronavirus, namely, human coronavirus OC43, human coronavirus HKU1,2 severe acute respiratory syndrome (SARS) coronavirus3,4 and Middle East respiratory syndrome (MERS) coronavirus,5 have also been found to infect human. The concept that human coronaviruses are generally associated with mild respiratory diseases was overturned by the identification in 2003 of SARS coronavirus, which causes a severe and highly lethal disease.3,4 Further investigation of the etiology of community-acquired pneumonia in an adult who was hospitalized in Hong Kong during the outbreak of SARS led to the identification of human coronavirus HKU1,2 which circulates commonly in human population causing respiratory tract illnesses worldwide.6,7,8 Although the mechanism by which SARS coronavirus and the newly identified MERS coronavirus cause severe respiratory diseases is not fully understood, their subversion of innate immunity is thought to contribute substantially to pathogenesis.4,9
Type I interferons (IFNs) are major effector cytokines in innate antiviral response. To induce IFN production, pathogen-associated molecular patterns such as viral double-stranded RNA are sensed by host pattern recognition receptors such as endosomal Toll-like receptor 3 and cytoplasmic RIG-I. The activation of these receptors transmits a signal to downstream kinases TBK1 and IKKε that form a functional complex with TRAF3 and TANK. Consequent phosphorylation of IRF3 and IRF7 transcription factors by these kinases leads ultimately to transcriptional activation of IFN promoters.10,11
Among the structural proteins encoded by SARS coronavirus, M protein is a glycoprotein with three N-terminal transmembrane domains named TM1, TM2 and TM3.12,13 We have previously reported that SARS coronavirus M protein suppresses type I IFN production potently by preventing the formation of functional TRAF3–TANK–TBK1/IKKε complex.14 This suppression of innate antiviral response by SARS coronavirus M protein represents one novel viral countermeasure against host innate immunity, which could play a role in SARS pathogenesis.9 However, it remains to be seen whether M proteins of other coronaviruses, which share 28%–41% identity in amino-acid sequence with SARS coronavirus M protein, might also exhibit IFN-antagonizing activity. Compared to SARS coronavirus, human coronavirus HKU1 is more frequently associated with less severe disease.6,7,8 It will therefore be of interest to determine whether human coronavirus HKU1 M protein would be able to suppress IFN production. On the other hand, the functional domain in SARS coronavirus M protein required and sufficient for IFN antagonism remains elusive.
In this study, we compared M proteins of SARS coronavirus and human coronavirus HKU1 for their ability to counteract IFN production. Furthermore, we dissected the structural domains in SARS coronavirus M protein and mapped the region that mediates the IFN-antagonizing activity.
Materials and methods
Plasmids and antibodies
pIFNβ-Luc reporter plasmid and RIG-I expression plasmid were obtained from Dr Takashi Fujita (Kyoto University, Kyoto, Japan).15 Expression vectors for TBK1, IRF3 and TRAF3 were generous gifts from Dr Genhong Cheng (University of California, Los Angeles, CA, USA).16,17 pISRE-Luc reporter construct was purchased from Clontech (Mountain View, CA, USA). MDA5, IKKε, MAVS and TANK expression vectors have been described.14
Monoclonal anti-FLAG (clone M2) and anti-α-tubulin (clone DM1A) antibodies were purchased from Sigma-Aldrich (St Louis, MO, USA). Anti-myc was bought from Santa Cruz (Dallas, TX, USA). Anti-V5 was from Life Technologies (Grand Island, NY, USA). Anti-GM130 was from BD Transduction (Lexington, KY, USA). Anti-calnexin was from Affinity Bioreagent (Golden, CO, USA).
Cell culture and transfection
HEK293 and HeLa cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine and 1% penicillin/streptomycin at 37 °C in a humidified atmosphere of 5% CO2. Cells were transfected with GeneJuice transfection reagent purchased from Novagen (Madison, WI, USA).
Protein analysis and reporter assay
Western blotting, immunoprecipitation and dual luciferase assay were performed as previously described.18,19 Relative luciferase activity (RLA) was derived by normalizing readings of firefly luciferase to those of Renilla luciferase. It was expressed in arbitrary units. All experiments were performed in triplicates and Student's t-test was used to assess statistically the differences between the indicated groups.
Confocal immunofluorescence microscopy
Confocal microscopy was performed on an LSM510 system (Carl Zeiss, Oberkochen, Germany) as described.20,21 Cell fixation was achieved with 1∶1 (v/v) acetone– methanol. Nuclei were counter-stained with 4′,6-diamidino-2-phenylindole.
Real-time reverse transcription polymerase chain reaction (RT-PCR)
Real-time RT-PCR was performed as described.22,23 Briefly, total RNA was extracted using RNAiso Plus reagent (Takara, Shiga, Japan) and cDNA was synthesized by Transcriptor First Strand cDNA kit (Roche, Indianapolis, IN, USA). Quantitation of target mRNA expression was achieved with the comparative Ct method.
Results
SARS coronavirus-specific inhibition of type I IFN production by M protein
M proteins from SARS coronavirus and human coronavirus HKU1 share 35% identity in amino-acid sequence (Figure 1). We have previously demonstrated the capability of SARS coronavirus M protein to antagonize type I IFN production.14 To determine whether human coronavirus HKU1 M protein could have similar activity, we made a side-by-side comparison of the two M proteins (Figure 2).
Amino-acid sequence alignment of M proteins from SARS coronavirus (SCV) and human coronavirus HKU1 (HKU1). Alignment was generated by Clustal ω (EMBL-EBI server (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The identity between the two proteins is 34.8%. Identical residues are indicated by asterisks (*). Strongly and weakly similar residues are highlighted by colons (:) and dots (.), respectively. SARS, severe acute respiratory syndrome.
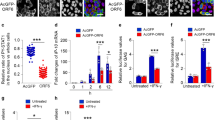
Suppression of IFN production by M protein is specific to SARS coronavirus. (a–h) Luciferase reporter assay. HEK293 cells in 24-well plates were transfected with the indicated reporter and expression plasmids. Fixed amount (50 ng) of expression plasmid for RIG-I, MDA5, MAVS and TBK1, fixed amount (100 ng) of reporter construct, as well as escalating amounts (100, 200 and 300 ng) of expression plasmids for SARS coronavirus (SCV) or human coronavirus HKU1 (HKU1) M protein were used. Cells were harvested for dual luciferase assay at 36 h post-transfection. Expression of V5-tagged M proteins was verified by western blotting (see right panel in a for one example). Results represent mean±s.d. derived from three independent experiments. P values by Student's t-test were all greater than 0.05 for the highlighted groups (#), indicating the lack of statistically significant difference. (i–k) Real-time RT-PCR analysis. Transfected HEK293 cells were harvested at 30 h post-transfection and real-time RT-PCR was performed to analyze IFN-β and GAPDH mRNAs. Relative levels of IFN-β mRNA expression were calculated from 2Ct(GAPDH)–Ct(IFN-β) and normalized to those recovered from cells receiving empty vector, which were set as 1. #P>0.05 by Student's t-test. GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IFN, interferon; RLA, relative luciferase activity; RT-PCR, reverse transcription polymerase chain reaction; SARS, severe acute respiratory syndrome; s.d., standard deviation.
We used two luciferase reporter constructs to measure the activation of IFN response in cultured cells. In plasmid pISRE-Luc, the expression of luciferase reporter was driven by IFN-stimulated response elements (ISRE), which are activated by type I IFNs and also bound to IRF3 and IRF7 transcription factors.24,25 For plasmid pIFNβ-Luc, reporter expression directly reflects the transcriptional activity of IFN-β promoter. To activate IFN production, we expressed RIG-I, MDA5, MAVS and TBK1 in HEK293 cells. RIG-I and MDA5 are sensors of viral double-stranded RNA.11,15 MAVS is a mitochondrial adaptor protein and TBK1 is the protein kinase that phosphorylates IRF3 and IRF7.1,11 These proteins represent three critical steps in the upstream, midstream and downstream of the intracellular signaling pathway that leads to IFN production. All proteins stimulated transcriptional activity driven by ISRE (Figure 1a–d) and IFN-β promoter (Figure 1e–h) in a dose-dependent manner. Consistent with our previous findings,14 SARS coronavirus M protein suppressed the stimulatory effect of RIG-I, MDA5, MAVS and TBK1 on ISRE and IFN-β promoter (Figure 1a–h). In stark contrast, human coronavirus HKU1 M protein had no influence on the activity of RIG-I, MDA5, MAVS or TBK1 (Figure 1a–h). Consistently, when we measured the steady-state levels of IFN-β transcript in transfected HEK293 cells by real-time RT-PCR, M protein of SARS coronavirus, but not of human coronavirus HKU1, inhibited the IFN-inducing activity of RIG-I, MDA5 and TBK1 in a dose-dependent fashion (Figure 1i–k). Thus, the suppression of IFN production by M protein is specific to SARS coronavirus.
N-linked glycosylation of SARS coronavirus M protein is not required for suppression of IFN production
We next sought to identify the molecular determinants in SARS coronavirus M protein that mediate virus-specific suppression of IFN production. N-linked glycosylation of viral structural proteins might affect their folding, stability, sorting and sensing by innate and adaptive immune systems.26 Since SARS coronavirus M protein undergoes N-linked glycosylation at a single site at the N terminus,12,27 we first investigated the requirement of this type of posttranslational modification for IFN antagonism. We constructed an N-linked glycosylation-defective mutant of SARS coronavirus M protein designated M-N4Q and interrogated its IFN-antagonizing activity. Since SARS coronavirus M protein and its M-N4Q mutant displayed equally potent activity to suppress RIG-I- and TBK-1-induced activation of IFN production (Figure 3), N-linked glycosylation is not influential in the suppression of IFN production by M protein.
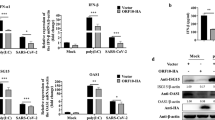
N-linked glycosylation is not required for suppression of IFN production by SARS coronavirus M protein. HEK293 cells in 24-well plates were cotransfected with fixed amount (50 ng) of an expression vector for RIG-I (a) or TBK1 (b), and with escalating amounts (100, 200 and 300 ng) of expression plasmid for either SARS coronavirus M or its N4Q mutant (M-N4Q) defective of N-linked glycosylation. Cells were harvested for dual luciferase assay at 36 h post-transfection. Data represent mean±s.d. of three independent experiments. IFN, interferon; RLA, relative luciferase activity; SARS, severe acute respiratory syndrome; s.d., standard deviation.
TM1 of SARS coronavirus M protein is required for IFN antagonism
SARS coronavirus M protein has three TMs (TM1: 1–38 amino acids; TM2: 51–69 residues; and TM3: 76–85 residues) and a cytoplasmic endodomain (86–221 amino acids).27 To determine which part of this protein is required for IFN antagonism, we made and expressed in HEK293 cells two truncated mutants designated M1 and M2 containing the TMs and the endodomain separately (Figure 4a and b). We next compared M and its mutants M1 and M2 for their ability to circumvent IFN induction by RIG-I and TBK1. Whereas M and M1 exhibited IFN-antagonizing activity of similar potency, M2 was unable to suppress IFN-β promoter activity (Figure 4b and c). Hence, the TMs are required for the suppressive effect of SARS coronavirus M protein on type I IFN production.
Suppression of IFN production by SARS coronavirus M protein is mediated by the N-terminal transmembrane domains. (a) Schematic diagram of SARS coronavirus M protein and its M1 and M2 truncated mutants. Amino-acid positions of the domains are indicated. (b, c) Luciferase reporter assay. HEK293 cells in 24-well plates were cotransfected with fixed amount (50 ng) of an expression vector for RIG-I or TBK1 and escalating amounts (100, 200 and 300 ng) of expression plasmids for M, M1 and M2 proteins. Shown above the bar plot in b is an example of western blot analysis of M, M1 and M2 proteins. #P>0.05. IFN, interferon; RLA, relative lucifearse activity; SARS, severe acute respiratory syndrome.
We further investigated which of the three TMs is required for IFN antagonism. Because the single TM was unstable and secreted out when expressed alone in cultured HEK293 cells (data not shown), we conjugated it to the endodomain, which was shown to be dispensable for IFN antagonism (Figure 4). The mutants named TM1′, TM2′ and TM3′, which respectively contain TM1 (amino acids 1–38), TM2 (amino acids 51–69) and TM3 (amino acids 76–85) fused to the endodomain, were constructed and expressed in HEK293 cells. All mutant proteins were stable and abundantly detected in the cell lysate (Figure 5a). Interestingly, only TM1′ was capable of inhibiting IFN-β promoter activity activated by RIG-I and TBK1. Neither TM2′ nor TM3′ exhibited IFN-antagonizing activity (Figure 5b and c). In further support of the importance of TM1, a chimeric M protein containing TM1 (residues 1–38) of SARS coronavirus and the cytoplasmic domain (87–223 residues) derived from human coronavirus HKU1 was equally active as SARS coronavirus M protein in the suppression of IFN production induced by RIG-I or TBK1, whereas another chimera carrying TM1 of human coronavirus HKU1 was totally inactive in this assay (Figure 6a-c). Taken together, because TM2, TM3 and endodomain are dispensable for IFN antagonism, only TM1 is required and probably sufficient for the suppression of IFN production.
Suppression of IFN production by SARS coronavirus M protein is mediated by TM1. (a) Western blot analysis of SARS coronavirus M protein and its TM1′, TM2′ and TM3′ mutants, a schematic diagram of which is shown above. (b, c) Luciferase reporter assay. HEK293 cells in 24-well plates were cotransfected with fixed amount (50 ng) of an expression vector for RIG-I or TBK1 and escalating amounts (100, 200 and 300 ng) of expression plasmids for M, TM1′, TM2′ and TM3′ proteins. #,# #P>0.05 by Student's t-test. IFN, interferon; RLA, relative luciferase activity; SARS, severe acute respiratory syndrome.
Influence of chimeric M proteins on IFN production. (a) Schematic diagram of chimeric M proteins. SCV M: SARS coronavirus M protein. HKU1 M: human coronavirus HKU1 M protein. (b, c) Luciferase reporter assay. HEK293 cells in 24-well plates were co-transfected with fixed amount (50 ng) of an expression vector for RIG-I or TBK1 and escalating amounts (100, 200 and 300 ng) of expression plasmids for SARS coronavirus M and chimeric M proteins chimera-1 and chimera-2. Shown above the bar plot in b is an example of western blot analysis of chimera-1 and chimera-2 proteins. #P>0.05 by Student's t-test. IFN, interferon; RLA, relative luciferase activity; SARS, severe acute respiratory syndrome.
Mechanism of TM1 suppression of IFN production
SARS coronavirus M protein suppresses IFN production by associating with and sequestering transducer proteins, thereby preventing the formation of functional TRAF3–TANK–TBK1/IKKε complex.14 If TM1 mediates IFN antagonism of SARS coronavirus M protein, it should be able to associate with transducer proteins and impede TRAF3 interaction with partners. In addition, because Golgi targeting would be the cause of transducer protein sequestration,14 TM1 should also be expected to be found in the Golgi apparatus.
Both SARS coronavirus M protein and the M1 mutant are known to localize predominantly to the Golgi apparatus.14,27 In agreement with this, SARS coronavirus M protein colocalized substantially with GM130 (Figure 7a–c), a marker of the Golgi complex.28 Interestingly, Golgi localization was also evident for TM1′ mutant (Figure 7g–i). Neither M nor TM1′ colocalized significantly with calnexin (Figure 7d–f and j–l), a marker of the endoplasmic reticulum.29 In contrast, TM2′ and TM3′ were largely concentrated in the endoplasmic reticulum. Their localization patterns were similar to that of calnexin, but distinct from that of GM130 (Figure 7m–x). In other words, TM2, TM3 or endodomain was not essential for Golgi targeting. Thus, TM1 is both required and probably sufficient for Golgi localization of SARS coronavirus M protein.
Subcellular localization of SARS coronavirus M protein and its mutants. HeLa cells were transfected with expression plasmids for V5-tagged SARS coronavirus M protein and its TM1′, TM2′ and TM3′ mutants. Cells were then stained for V5 and a marker protein. Nuclear morphology (blue) was visualized with DAPI. Specific fluorescent signals from V5 and the marker were then merged. Transfected cells in the merged panels are highlighted by arrows. Colocalization appeared yellow. GM130 and calnexin serve as markers for the Golgi complex and the ER, respectively. All panels shown are representative of the results of triplicate experiments. Whereas significant colocalization of M and TM1′ with GM130 was seen in 83% and 87% of 100 transfected cells, TM2′ and TM3′ colocalized with calnexin significantly in 66% and 73% of 100 transfected cells. Bar=20 µm. DAPI, 4′,6-diamidino-2-phenylindole; ER, endoplasmic reticulum; IFN, interferon; SARS, severe acute respiratory syndrome.
On the other hand, reciprocal immunoprecipitation and immunoblotting experiments indicated the association of TM1′ with RIG-I, TRAF3, TBK1 and IKKε (Figure 8a, lanes 1–4). Neither TM2′ nor TM3′ had the same property (Figure 8a, lanes 5–12). In addition, the interaction of TRAF3 with TBK1, IKKε and TANK was inhibited in the presence of TM1′ (Figure 8b, lanes 1–6), but not TM2′ or TM3′ (Figure 8b, lanes 7–10). In light of this, TM1 is both required and probably sufficient for interacting with TRAF3 and other transducer proteins and thereby preventing TRAF3 from engaging its partners such as TBK1, IKKε and TANK. Indeed, both SARS coronavirus M protein and its TM1′ mutant colocalized with TRAF3 (Figure 9a–f). However, human coronavirus HKU1 M protein also localized to the Golgi apparatus (data not shown) and colocalized with TRAF3 (Figure 9g–i), but did not impede the interaction of TRAF3 with TANK, TBK1 or IKKε (data not shown). Plausibly, Golgi localization and interaction with TRAF3 are required but not sufficient for suppression of IRF3 activation.
Mechanism of TM1 suppression of IFN production. (a) Interaction of TM1 with RIG-I, TRAF3, TBK1 and IKKε. HEK293 cells were transfected with the indicated plasmids expressing tagged proteins. Reciprocal immunoprecipitation (IP) and western blotting were performed with anti-FLAG (α-FLAG) and anti-V5 (α-V5) antibodies. (b) Suppression of TRAF3–TANK–TBK1/IKKε complex formation by TM1′, but not TM2′ or TM3′. Co-immunoprecipitation was performed with anti-FLAG and lysates of transfected HEK293 cells. The input lysates (5%) were also probed for TBK1/IKKε/TANK, TRAF3 and TM1′/TM2′/TM3′ by western blotting. IFN, interferon.
Colocalization of SARS coronavirus M protein and its TM1′ mutant with TRAF3. HeLa cells were transfected with plasmids expressing TRAF3 and V5-tagged SARS coronavirus M protein (SCV M), its TM1′ mutant or human coronavirus HKU1 M protein (HKU1 M). Cells were then stained for M and TRAF3. Nuclear morphology (blue) was visualized with DAPI. Specific fluorescent signals from V5 and TRAF3 were then merged. Transfected cells in the merged panels are highlighted by arrows. Colocalization appeared yellow. All panels shown are representative of the results of triplicate experiments. Bar=20 µm. DAPI, 4′,6-diamidino-2-phenylindole; IFN, interferon; SARS, severe acute respiratory syndrome.
Discussion
In this study, we showed that SARS coronavirus M protein suppressed the production of type I IFNs in a virus-specific manner and it did not share IFN-antagonizing property with M protein of human coronavirus HKU1. IFN antagonism of SARS coronavirus M protein was mediated by N-terminal TM1 (amino acids 1–38), which targets M protein to the Golgi complex and associates with TRAF3 to prevent it from interacting with TANK, TBK1 and IKKε. Our findings provide additional molecular details for suppression of type I IFN production by SARS coronavirus M protein.
The inability of human coronavirus HKU1 M protein to suppress IFN production was not too surprising. Although the two full M proteins of SARS coronavirus and human coronavirus HKU1 share 35% identity, only 26% of the amino-acid residues in the TM1 region of the two proteins are identical. This difference in the TM1 region might be critical in governing the inhibition of TRAF3. SARS coronavirus is known to suppress innate antiviral response at multiple levels leading to a severe disease.4,9 This might be similar to the emerging MERS coronavirus.30,31 In contrast, human coronavirus HKU1 is commonly found in human population and is associated with less severe respiratory illnesses.6,7,8 Accordingly, it might not have the same ability to evade innate immunity. Viral proteins are not well characterized and IFN-antagonizing proteins have not been identified in human coronavirus HKU1.32 It remains to be seen whether the distinct properties of M proteins in the suppression of IFN production could account at least partly for the severity of disease caused by SARS coronavirus and human coronavirus HKU1.
In one model to explain this difference, SARS coronavirus and MERS coronavirus are not yet adapted to the new environment in human,4,9,33 whereas the well-adapted human coronavirus HKU1 and the other human coronaviruses such as OC43 and 229E have lost some of their immunosuppressive tools during evolution. To test this idea, the coronaviruses in the two groups should be compared more thoroughly for their abilities to induce IFNs and to suppress IFN production and action. Particularly, the IFN-antagonizing property of M proteins from more coronaviruses should be assessed in parallel. Although attempts have been made to culture the virus in primary human ciliated airway epithelial cells and type II alveolar epithelial cells,34,35,36 clinical isolates of human coronavirus HKU1 remain unculturable in most laboratories. This hampers further comparative study in cultured cells. To remedy this, the dynamics of IFN induction should be analyzed in human subjects naturally infected with human coronavirus HKU1.
We not only defined a minimal TM1 domain in SARS coronavirus M protein that mediates suppression of IFN production, but also provided new mechanistic details for this suppression. Although TM1 alone is unstable and secreted out, it is stable and functional when conjugated to TM2–TM3 or endodomain of SARS coronavirus M protein or of human coronavirus M protein (data not shown). TM1 contains the critical determinants for Golgi localization, for association with RIG-I, TRAF3, TBK1 and IKKε, and for prevention of TRAF3 from interacting with TANK, TBK1 and IKKε. Our results are compatible with the notion that TM1 targets SARS coronavirus M protein and its associated cellular proteins to the Golgi apparatus. It will be of importance to clarify exactly how this might contribute to the suppression of IRF3 activation.
Notably, human coronavirus HKU1 M protein also targets the Golgi complex, interacts with TRAF3, but does not suppress IFN production. Hence, Golgi localization and interaction with TRAF3 are required but not sufficient for IFN antagonism. Mechanistically, the perturbation of the interaction of TRAF3 with TANK, TBK1 and IKKε by TM1 of SARS coronavirus M protein is more critical in the suppression of IRF3 activation. This could plausibly be mediated by pre-occupation and steric hindrance, but further investigations are required to provide additional support to this model. For example, it will be of interest to see whether artificial fusion of TM1 to TRAF3 might impede its binding with TBK1 and IKKε or inhibit its activation of IFN production. Mapping and comparing the TM1- and TBK1-interacting domains in TRAF3 would also shed light on how the binding of TRAF3 with TM1 affects the binding with TBK1. More importantly, a thorough comparison of the TM1 regions of SARS coronavirus and human coronavirus HKU1 will define the critical residues for the suppression of TRAF3 interaction with downstream transducers. Nevertheless, this model of suppression by sequestration and pre-occupation represents a new mechanism for immune evasion, which might also be used by other viral and cellular proteins such as microtubule-associated TRAF3-binding protein MIP-T3, which localizes to the centrosome and cilia and also impedes the formation of functional TRAF3-containing complex.37 Furthermore, our definition of a small TM1 domain that mediates immune evasion will pave the way for rational design and development of new immunosuppressive agents. In this regard, both peptide mimetics and recombinant proteins that mimic the action of TM1 might prove useful.
References
Randall RE, Goodbourn S . Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol 2008; 89: 1–47.
Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia, J Virol 2005; 79: 884–895.
Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003; 361: 1319–1325.
Cheng VC, Lau SK, Woo PC, Yuen KY . Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev 2007; 20: 660–694.
Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA . Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012; 367: 1814–1820.
Lau SK, Woo PC, Yip CC, Tse H, Tsoi HW, Cheng VC et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol 2006; 44: 2063–2071.
Jevšnik M, Uršič T, Zigon N, Lusa L, Krivec U, Petrovec M . Coronavirus infections in hospitalized pediatric patients with acute respiratory tract disease. BMC Infect Dis 2012; 12: 365.
Woo PC, Lau SK, Yip CC, Huang Y, Yuen KY . More and more coronaviruses: human coronavirus HKU1. Viruses 2009; 1: 57–71.
Totura AL, Baric RS . SARS coronavirus pathogenesis: host innate immune responses and viral antagonism of interferon. Curr Opin Virol 2012; 2: 264–275.
Moresco EM, LaVine D, Beutler B . Toll-like receptors. Curr Biol 2011; 21: R488–R493.
Goubau D, Deddouche S, Reis E, Sousa C . Cytosolic sensing of viruses. Immunity 2013; 38: 855–869.
Oostra M, de Haan CA, de Groot RJ, Rottier PJ . Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J Virol 2006; 80: 2326–2336.
Siu YL, Teoh KT, Lo J, Chan CM, Kien F, Escriou N et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol 2008; 82: 11318–11330.
Siu KL, Kok KH, Ng MH, Poon VK, Yuen KY, Zheng BJ et al. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3⋅TANK⋅TBK1/IKKε complex. J Biol Chem 2009; 284: 16202–16209.
Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 2004; 5: 730–737.
Guo B, Cheng G . Modulation of the interferon antiviral response by the TBK1/IKKi adaptor protein TANK. J Biol Chem 2007; 282: 11817–11826.
Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T et al. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 2002; 17: 251–263.
Kok KH, Lui PY, Ng MH, Siu KL, Au SW, Jin DY . The double-stranded RNA-binding protein PACT functions as a cellular activator of RIG-I to facilitate innate antiviral response. Cell Host Microbe 2011; 9: 299–309.
Siu KL, Chan CP, Chan C, Zheng BJ, Jin DY . Severe acute respiratory syndrome coronavirus nucleocapsid protein does not modulate transcription of human FGL2 gene. J Gen Virol 2009; 90: 2107–2113.
Ching YP, Chan SF, Jeang KT, Jin DY . Retroviral oncoprotein Tax targets coiled-coil centrosomal protein TAX1BP2 to induce centrosome overduplication. Nat Cell Biol 2006; 8: 717–724.
Chin KT, Chun AC, Ching YP, Jeang KT, Jin DY . Human T-cell leukemia virus oncoprotein Tax represses nuclear receptor-dependent transcription by targeting coactivator TAX1BP1. Cancer Res 2007; 67: 1072–1081.
Chan CP, Kok KH, Tang HM, Wong CM, Jin DY . Internal ribosome entry site-mediated translational regulation of ATF4 splice variant in mammalian unfolded protein response. Biochim Biophys Acta 2013; 1833: 2165–2175.
Tang HM, Gao WW, Chan CP, Siu YT, Wong CM, Kok KH et al. LKB1 tumor suppressor and salt-inducible kinases negatively regulate human T-cell leukemia virus type 1 transcription. Retrovirology 2013; 10: 40.
Schafer SL, Lin R, Moore PA, Hiscott J, Pitha PM . Regulation of type I interferon gene expression by interferon regulatory factor-3. J Biol Chem 1998; 273: 2714–2720.
Zhang L, Pagano JS . Interferon regulatory factor 7 is induced by Epstein–Barr virus latent membrane protein 1. J Virol 2000; 74: 1061–1068.
Vigerust DJ, Shepherd VL . Virus glycosylation: role in virulence and immune interactions. Trends Microbiol 2007; 15: 211–218.
Voss D, Pfefferle S, Drosten C, Stevermann L, Traggiai E, Lanzavecchia A et al. Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein. Virol J 2009; 6: 79.
Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P et al. Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol 1995; 131: 1715–1726.
Rajagopalan S, Xu Y, Brenner MB . Retention of unassembled components of integral membrane proteins by calnexin. Science 1994; 263: 387–390.
Niemeyer D, Zillinger T, Muth D, Zielecki F, Horvath G, Suliman T et al. Middle East respiratory syndrome coronavirus accessory protein 4a is a type I interferon antagonist. J Virol 2013; 87: 12489–12495.
Siu KL, Yeung ML, Kok KH, Yuen KS, Kew C, Lui PY et al. Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in innate antiviral response. J Virol 2014; 88: in press.
Siu KL, Chan CP, Woo PC, Jin DY . Comparative analysis of the activation of unfolded protein response by spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus HKU1. Cell Biosci 2014; 4: 3.
Chan JF, Lau SK, Woo PC . The emerging novel Middle East respiratory syndrome coronavirus: The “knowns” and “unknowns”. J Formos Med Assoc 2013; 112: 372–381.
Pyrc K, Sims AC, Dijkman R, Jebbink M, Long C, Deming D et al. Culturing the unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J Virol 2010; 84: 11255–11263.
Dominguez SR, Travanty EA, Qian Z, Mason RJ . Human coronavirus HKU1 infection of primary human type II alveolar epithelial cells: cytopathic effects and innate immune response. PLoS ONE 2013; 8: e70129.
Dijkman R, Jebbink MF, Koekkoek SM, Deijs M, Jónsdóttir HR, Molenkamp R et al. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J Virol 2013; 87: 6081–6090.
Ng MHJ, Ho TH, Kok KH, Siu KL, Li J, Jin DY . MIP-T3 is a negative regulator of innate type I interferon response. J Immunol 2011; 187: 6473–6482.
Acknowledgements
We thank Genhong Cheng and Takashi Fujita for reagents; and Chun Kew, Pak-Yin Lui, Vincent Hei-Man Tang and Kit-San Yuen for critical reading of manuscript. This work was supported by Hong Kong Health and Medical Research Fund (10091192) and Hong Kong Research Grants Council (HKU1/CRF/11G).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Siu, KL., Chan, CP., Kok, KH. et al. Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cell Mol Immunol 11, 141–149 (2014). https://doi.org/10.1038/cmi.2013.61
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cmi.2013.61
Keywords
This article is cited by
-
Innate immune sensing of coronavirus and viral evasion strategies
Experimental & Molecular Medicine (2021)
-
Viral pathogen-induced mechanisms to antagonize mammalian interferon (IFN) signaling pathway
Cellular and Molecular Life Sciences (2021)
-
Allo-priming as a universal anti-viral vaccine: protecting elderly from current COVID-19 and any future unknown viral outbreak
Journal of Translational Medicine (2020)
-
PCR array profiling of antiviral genes in human embryonic kidney cells expressing human coronavirus OC43 structural and accessory proteins
Archives of Virology (2018)
-
Pathogenic human coronavirus infections: causes and consequences of cytokine storm and immunopathology
Seminars in Immunopathology (2017)