Abstract
Chromosome aneuploidy is a major cause of pregnancy loss, abnormal pregnancy and live births following both natural conception and in vitro fertilisation (IVF) and increases exponentially with maternal age in the decade preceding the menopause. Molecular genetic analysis following natural conception and spontaneous miscarriage demonstrates that trisomies arise mainly in female meiosis and particularly in the first meiotic division. Here, we studied copy number gains and losses for all chromosomes in the two by-products of female meiosis, the first and second polar bodies, and the corresponding zygotes in women of advanced maternal age undergoing IVF, using microarray comparative genomic hybridisation (array CGH). Analysis of the segregation patterns underlying the copy number changes reveals that premature predivision of chromatids rather than non-disjunction of whole chromosomes causes almost all errors in the first meiotic division and unlike natural conception, over half of aneuploidies result from errors in the second meiotic division. Furthermore, most abnormal zygotes had multiple aneuploidies. These differences in the aetiology of aneuploidy in IVF compared with natural conception may indicate a role for ovarian stimulation in perturbing meiosis in ageing oocytes.
Similar content being viewed by others
Introduction
Chromosome aneuploidy is a major cause of pregnancy loss, abnormal pregnancy and live births following both natural conception and in vitro fertilisation (IVF) and increases exponentially with maternal age in the decade preceding the menopause.1, 2 The classic textbook mechanism for aneuploidy arising in meiosis is non-disjunction of whole chromosomes or sister chromatids in the first (meiosis I) and second meiotic divisions (meiosis II), respectively.3 Molecular genetic analysis of human trisomies following natural conception has demonstrated that most arise in female meiosis and heterozygosity of polymorphic markers adjacent to the centromere has been taken as evidence that most of these arise by non-disjunction in meiosis I, though there is some variation between chromosomes.1, 4 Karyotyping of human metaphase II oocytes from women having IVF, however, demonstrated that at least some trisomies are caused by malsegregation not of whole chromosomes but of single chromatids in meiosis I. This presumably results from premature predivision of the whole chromosomes into sister chromatids, which then segregate at random.5 Furthermore, this interpretation is consistent with fluorescence in situ hybridisation (FISH) analyses of aneuploidy for five to eight mainly acrocentric chromosomes in the two by-products of female meiosis, the first (PB1) and second polar bodies (PB2), that is, the majority of errors in PB1 were detected as one or three hybridisation signal(s) for a particular chromosome instead of the normal two.6, 7
The incidence of chromosome aneuploidy in human preimplantation embryos following IVF, particularly in women of advanced maternal age, is high and is assumed to contribute to the rapid decline in pregnancy and live birth rates. Hence, preimplantation genetic screening (PGS) for aneuploidy by either polar body or cleavage stage biopsy of single cells has been widely practiced. However, a number of randomised clinical trials have now demonstrated that FISH analysis for a limited number of chromosomes in single cells biopsied from cleavage stage embryos, has either no effect or reduces live birth rates when applied to all women in their late 30s or early 40s.8 The European Society of Human Reproduction and Embryology therefore set up a PGS task force, which decided to organise a clinical pilot study to investigate the feasibility and accuracy of using microarray-based comparative genomic hybridisation (array CGH) for copy number analysis of all 23 different chromosome pairs in both polar bodies. This enables the identification of aneuploidies arising specifically in female meiosis.9 The main advantages of this approach are that as the polar bodies do not form part of the developing embryo, removing them for analysis is less invasive. Also, polar body analysis avoids diagnostic errors resulting from chromosome mosaicism caused by malsegregation of chromosomes or chromosome loss in the early mitotic divisions, which is relatively common in cleavage stage embryos.10, 11
The clinical outcome and technical aspects of this pilot study have been published in detail elsewhere.12, 13 Briefly, in a series of women having aneuploidy testing mainly because of advanced maternal age (mean age 40 years), array CGH results were obtained for both polar bodies in a high proportion of the oocytes examined (86%) of which nearly three quarters (72%) had one or more aneuploidies in either one or both polar bodies, predicting aneuploidy in the corresponding fertilised oocytes (zygotes). In these cases and a small number with euploid polar bodies, the corresponding zygotes were then analysed blind to determine the rate of concordance for their overall euploid/aneuploid status. Eliminating a small number of results from zygotes in which contamination with maternal DNA had clearly occured, the concordance rate was high (94%). Furthermore, the results were available within 12–13 h enabling aneuploidy testing in countries like Germany, which at the time of the study did not allow genetic testing of the embryo. Ongoing clinical pregnancy rates of 17% per cycle and 30% per embryo transfer are encouraging. However, a multicentre randomised trial is now underway in women of advanced maternal age to determine whether this less invasive and comprehensive testing of aneuploidy improves the clinical outcome for these patients.
Here we have examined the copy number changes resulting from meiotic errors, in the three products of female meiosis, that is, both polar bodies and the corresponding zygote, to infer the mechanism causing the errors. Thus, we have been able to distinguish non-disjunction from premature predivision of sister chromatids as the cause of aneuploidy in the corresponding zygotes. The results of this analysis demonstrate that almost all meiosis I errors for all chromosomes are caused by premature predivision of sister chromatids. As would be expected by the random segregation of single chromatids at meiosis II to either the PB2 or the zygote, however, a significant proportion of meiosis I errors did not result in aneuploidy in the zygote. Overall, therefore, there were more meiosis II-derived maternal aneuploidies in the zygote. Furthermore, over half of the zygotes examined had multiple aneuploidies. These differences in the aetiology of aneuploidy in IVF compared with natural conception may indicate a role for ovarian stimulation in perturbing meiosis in ageing oocytes.
Materials and methods
A total of 34 couples undergoing IVF with PGS for aneuploidy, either because of advanced maternal age (n=31) or a maternal balanced translocation (n=3), consented to polar body biopsy and aneuploidy testing by array CGH. The average maternal age was exactly 40.0 years (range 33–44). Intracytoplasmic sperm injection of mature metaphase II oocytes was carried out ∼3–8 h after egg collection and PB1 and PB2 biopsied simultaneously from all normally fertilised oocytes between 6 and 9 h later. The two polar bodies were then processed separately. Each polar body was washed and placed in a small volume of PBS in a PCR tube, the whole genome amplified by PCR and the DNA labelled for array CGH (24Sure, BlueGnome, Cambridge, UK) as described in detail elsewhere.12, 13, 14 Following array CGH, each polar body was analysed for gains and losses and the euploid/aneuploid status of the corresponding zygote predicted. This process took between 12 and 13 h. If copy number gains and/or losses were detected by array CGH, the zona pellucida was removed from the corresponding presumed aneuploid zygote (still at the 1-cell stage before the first mitotic division) and after washing and collecting in PBS, sent to another laboratory, blinded to the polar body results, for array CGH. In a small number of cases, presumed euploid zygotes of poor morphology, which would not have been considered for transfer, were also analysed.
Segregation pattern analysis
To examine the incidence of aneuploidy in the two meiotic divisions and to distinguish the two possible mechanisms, 105 sets of complete array CGH data, including PB1, PB2 and the corresponding zygote, were available for analysis. Data from zygotes in which there was clear evidence of contamination or any other technical issues affecting the array CGH results were excluded. Although gain or loss of whole chromosomes versus single chromatids in PB1 was apparent in some cases by the extent of the altered ratio following array CGH (especially where examples of both were present in the same plot), this was not systematically analysed and not considered completely reliable. Alternatively, however, whole chromosomes versus chromatid errors in meiosis I can be distinguished by analysing the segregation pattern in all three products of meiosis. Therefore, for each chromosome copy number gain (G) or loss (L) detected in either of the two polar bodies or zygote, the corresponding gain, loss or (N) normal copy number in the other two meiotic products was recorded as a three letter code representing the copy number in PB1/PB2/zygote. Of the possible permutations, there are only five possible patterns for both gains and losses in the zygote arising from errors in maternal meiosis I by either whole chromosome non-disjunction or premature predivision of sister chromatids or in meiosis II (Figure 1; Table 1). In addition, there are two patterns indicating the absence of an error in either female meitoic division, but either introduced by the fertilising sperm, or caused by early chromosome loss before the first mitotic division.
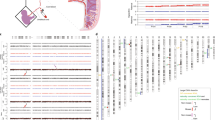
Diagramatic representation of copy number segregation patterns resulting from normal disjunction, non-disjunction of whole chromosomes and premature predivision of chromatids in a malsegregating chromosome. Note that not all possible segregation patterns are represented and reciprocal patterns are also possible. In each panel, the first polar body (PB1) is on the left and the seond polar body (PB2) is on the right with part of the corresponding zygote below. Maternal chromosomes (red); paternal chromosomes (blue). The segregation pattern is displayed on the right as gain (G), loss (L) or normal (N) copy number for PB1/PB2/Zygote.
Results
Excluding the chromosomes involved in the three translocation cases, and 25 de novo structural abnormalities, 2376 chromosomes were analysed in the 105 array CGH data sets, which included PB1, PB2 and the corresponding zygote (Figure 2). Of these, 2023 had a normal segregation pattern (NNN) and 353 (15%) had copy number gains or losses in either one or both polar bodies and/or the zygote. In all, 275 (78%) of these could be categorised according to the predicted segregation patterns for errors in meiosis or in the zygote (Table 2). The other 78 did not correspond to any of the recognised patterns and were almost all copy number changes in only one of the polar bodies (see Supplementary data).
Array CGH ratio plots for the first and second polar bodies (PB1 and PB2) and corresponding zygote in which there are four errors in meiosis I and five in meiosis II resulting in six gains (trisomies) and one loss (monosomy) in the corresponding zygote. The segregation patterns of gains (G), losses (L) and normal copy numbers (N) indicate meiosis I nondisjunction (LGG) for chromosome 18, premature predivision (LNG and GNL) for chromosomes 9 and 13, respectively, and an error balanced in meiosis II (LGN) for chromosome 20. The other four meiosis II errors all resulted in gains in the zygote (NLG) for chromosomes 4 (arrows), 10, 15 and 21. The two horizontal lines either side of log2 ratio 0.0 represent the 95% confidence interval for normal copy number.
Of 125 meiosis I errors, detected by gain or loss in PB1, 77 (62%) resulted in aneuploidy in the corresponding zygote with normal copy number in PB2, as would be expected by malsegregation of sister chromatids following premature predivision (Table 2). While another 48 (38%) meiosis I errors were combined with a balancing gain or loss in PB2, resulting in disomy in the corresponding zygote. Again, this is consistent with predivision and random segregation of single chromatids in meiosis II. Only four (3%) meiosis I errors had the expected segregation patterns for non-disjunction resulting in copy number change in PB2 and aneuploidy in the corresponding zygotes. In addition, there were 102 meiosis II errors resulting in aneuploidies in the corresponding zygotes, in which copy number in PB1 was normal, and 48 presumed paternal aneuploidies or anaphase losses, only present in the zygote. Therefore, although the incidence of errors occuring at meiosis I (detected by abnormal copy number in PB1) was more frequent than those occuring at meiosis II (detected by normal copy number in PB1 but gain or loss in PB2), significantly more net gains and losses in the aneuploid zygotes were caused by errors in meiosis II (Table 3). Furthermore, there was a trend towards losses from the polar bodies in both divisions. There was also a higher incidence of losses compared with gains in aneuploidies only present in the zygote. This suggests that early chromosome loss may occur possibly as a result of anaphase lag before the first mitotic division.
Aneuploidies arising from errors in maternal meiosis were spread across all chromosomes, with the exception of chromosome 3, with a trend towards an increased incidence for the smaller and acrocentric chromosomes (Table 2; Figure 3). The most frequent aneuploidies were for chromosomes 16 and 22, followed by 21, 19, 11 and 15. Although it varied between chromosomes, overall gains and losses were approximately equally represented. Over half (58%) aneuploid zygotes had multiple maternally derived aneuploidies (range 2–7; Figure 4). Furthermore, the incidence of multiple aneuploidies was strongly age dependent reaching 87% of aneuploid zygotes in women between 43 and 45 years of age (Table 4).
Histogram of the number of aneuploidies resulting from maternal meiotic and other errors per oocyte (note that in some cases no maternal aneuploidies were present either because a few zygotes with euploid polar bodies were included or there was only a combination of MI errors balanced in MII and/or segregation patterns that could not be categorised).
Discussion
Our observations reveal a pattern of multiple meiotic errors, typically caused by predivision of chromatids and predominantly arising at meiosis II, in the aneuploid zygotes of women of advanced maternal age undergoing IVF. Furthermore, these data are unique because, unlike spontaneous miscarriages, it is not influenced by the viability of the conceptus and is thus representative of the range of aneuploidies at conception, including those that may be lethal at preimplantation stages of development. The pattern we observed contrasts sharply with the patterns of aneuploidy that have been reported for miscarriages or live births. Most notably, although spontaneous miscarriage is often associated with karyotype abnormalities, it is uncommon to find two or more aneuploidies even when uncultured samples from the first trimester are analysed by array CGH.15 Nevertheless, the incidence of double trisomies, although relatively rare, is strongly maternal age dependent following natural conception and includes a wide range of chromosomes though never chromosomes 1 or 19.16 In our data, only a single maternal aneuploidy for chromosome 1 (monosomy) was detected but there were 10 zygotes with trisomy 19 of which 6 had one or more other aneuploidies (Figure 3; Supplementary data).
In all, 70% of pregnancies and live births affected by trisomy 21 (Down syndrome) following natural conception, originate in female meiosis I (as assessed by heterozygosity of polymorphic maternal markers close to the centromere).2 Of 16 chromosome, 21 gains detected in zygotes here, only 4 originated in meiosis I, 9 in meiosis II and 3 were presumed to be of paternal origin (Table 2). Furthermore, the pattern is similar in aneuploid zygotes with only a single maternal trisomy for one of the acrocentric chromosomes known to be compatible with implantation and clinical pregnancy (see Supplementary data). Interestingly, a recent study of a large cohort of familes with a live born infant with trisomy 21 demonstrated a reduced meiosis I/meiosis II ratio in women <19 and ≥40 years of age compared with those of intermediate ages.17 However, it is important to recognise that, like the data from trisomies, we can only infer the stage at which errors arise indirectly when malsegregation occurs. Many of the errors scored here as meiosis II errors almost certainly originate in meiosis I since premature separation – either of homolgues or of sister chromatids – will not always lead to a segregation error. Thus, at least a proportion of meiosis II errors may be undetected meiosis I errors.
In contrast, trisomy 8 in spontaneous miscarriages is generally caused by postzygotic mitotic duplication following normal conception4 whereas here, there are five meiosis I errors, four of which are balanced in meiosis II and two meiosis II errors. However, at least one homogeneously affected fetus with trisomy 8 has been reported to be of meiosis I origin though the parental origin could not be confirmed.18 Although the sample size is limited, the observed distribution of gains does not match closely the frequencies of trisomies observed among all recognised pregnancies (Figure 2). In particular, the high frequency of gains for chromsomes 11 and 19, the absence of trisomy 7 and the single trisomy 14 are unusual findings. But the absence of gain or loss for chromosome 3 is not unusual. Also, in miscarriages, monosomy X is as frequent as the most frequent trisomy, trisomy 16, whereas here, there are very few aneuploidies for the sex chromosomes. This suggests that monosomy X is mostly caused by postzygotic losses perhaps through anaphase lag. Alternatively, it might simply reflect the difficulty of detecting copy number changes in the sex chromosomes accurately against sex-matched or -unmatched control DNA as is the current practice in the array CGH method used.
Of the 78 (22%) copy number changes with unrecognised segregation patterns, 73 were changes in only one of the polar bodies (see Supplementary data). Assuming the normal number of whole chromosomes and sister chromatids were present in these oocytes before meiosis, isolated gain or loss in either PB1 or PB2 is not theoretically possible. Some of these apparent copy number changes may therefore be technical artifacts (false positives). However, because they were frequently associated with other aneuploidies in the same zygote, the high concordance rate (94%) at the level of the overall euploid/aneuploid status of the zygote was not seriously affected.12 Alternatively, another likely explanation is the failure to detect the corresponding gain or loss in one of the other two meiotic products (false negatives) possibly because of technical artifacts again or contamination with maternal DNA.12 In this case, the actual segregation patterns for these changes would follow the same distribution as those with the recognised patterns (see Supplementary data). Finally, it is also possible that some losses from polar bodies do not cause corresponding gains in the other polar body or zygote because of chromosome loss through anaphase lag.
The cause of aneuploidy in the human female is still not fully understood. Apart from the well-known association with advanced maternal age, the only other significant correlation is with a reduced number or altered distribution of recombination events in trisomies, which varies between chromosomes. It has been proposed, therefore, that factors at three distinct stages in the development of the mature female gamete (or oocyte) may be involved, beginning prenatally with factors influencing recombination patterns before arrest in prophase of metaphase I, age-related changes during follicle recruitment and growth and, resumption and completion of meiosis before ovulation and fertilisation, which may occur several decades later.19 In mice, gene targeting has shown that reduced or null expression of several cohesin genes involved in the synaptonemal complex responsible for cohesion of homologous chromosomes and sister chromatids, as well as genes involved in recombination, result in meiotic disruption or arrest.19 However, no quantitative differences in expression have been detected in cohesins in aneuploid human oocytes.20 Interestingly, targeting of cdc20, the activating subunit of the anaphase-promoting complex, caused various abnormalities with striking similarities to human preimplantation development, including misalignment of chromosomes on the meiosis I spindle, poor development and arrest of embryos before the blastocyst stage and a high incidence of chromosome lagging and aneuploidy.21
An alternative explanation for the maternal age-related increase in trisomies, the oocyte mosaic selection model, has recently been proposed.22 According to this model, there is a progressive selection of trisomic oogonia with age, which then undergo an obligate secondary non-disjunction in meiosis I. Although several possible segregation patterns may occur in secondary non-disjunction, because of the extra chromatids involved, the principal patterns resulting in trisomy in the zygote would be ‘GNG’ or ‘NGG’ and only one out of 353 copy number changes had this pattern (see Supplementary Information). There is therefore no evidence for gonadal mosaicism in our data. However, in some patients suffering repeated pregnancies with trisomy 21, for example, there is evidence from FISH studies of gonadal mosacism and an extra whole chromosome 21 in some oocytes.7, 23 In these cases, altered segregation patterns would presumably be detected.
The relatively high incidence of predivision observed here within individual oocytes, across all chromosomes, suggests that failure of cohesion is a major factor, at least in oocytes collected from women of advanced maternal age undergoing IVF. This may, therefore, indicate a role for ovarian stimulation in perturbing meiosis in ageing oocytes particularly as women of advanced maternal age often require higher doses of stimulatory drugs. In the mouse, the addition of follicle-stimulating hormone during in vitro maturation of oocytes, or simply IVF itself, increased the incidence of aneuploidy in meiosis I.24, 25 Further work comparing the incidence and pattern of meiotic errors following different stimulation regimes, including mild stimulation and natural cycle IVF, as well as analysis of the meiotic origin of aneuploid IVF pregnancies may identify improved clinical strategies to reduce the incidence of aneuploidy in these women and perhaps increase the success rates of IVF.
References
Hassold T, Hall H, Hunt P : The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet 2007; 16: R203–R208.
Spandorfer SD, Davis OK, Barmat LI, Chung PH, Rosenwaks Z : Relationship between maternal age and aneuploidy in in vitro fertilization pregnancy loss. Fertil Steril 2004; 81: 1265–1269.
Gardner RJM, Sutherland GR : Chromosome Abnormalities and Genetic Counseling. New York, USA: Oxford University Press, 2004.
Nicolaidis P, Petersen MB : Origin and mechanisms of non-disjunction in human autosomal trisomies. Hum Reprod 1998; 13: 313–319.
Angell RR : Predivision in human oocytes at meiosis I: a mechanism for trisomy formation in man. Hum Genet 1991; 86: 383–387.
Kuliev A, Verlinsky Y : Meiotic and mitotic nondisjunction: lessons from preimplantation genetic diagnosis. Hum Reprod Update 2004; 10: 401–407.
Cupisti S, Conn CM, Fragouli E et al: Sequential FISH analysis of oocytes and polar bodies reveals aneuploidy mechanisms. Prenat Diagn 2003; 23: 663–668.
Harper J, Coonen E, De Rycke M et al: What next for preimplantation genetic screening (PGS)? A position statement from the ESHRE PGD Consortium Steering Committee. Hum Reprod 2010; 25: 821–823.
Geraedts J, Collins J, Gianaroli L et al: What next for preimplantation genetic screening? A polar body approach!. Hum Reprod 2010; 25: 575–577.
Munne S, Weier HU, Grifo J, Cohen J : Chromosome mosaicism in human embryos. Biol Reprod 1994; 51: 373–379.
Delhanty JD, Harper JC, Ao A, Handyside AH, Winston RM : Multicolour FISH detects frequent chromosomal mosaicism and chaotic division in normal preimplantation embryos from fertile patients. Hum Genet 1997; 99: 755–760.
Geraedts J, Montag M, Magli MC et al: Polar body array CGH for prediction of the status of the corresponding oocyte. Part I: clinical results. Hum Reprod 2011; 26: 3173–3180.
Magli MC, Montag M, Koster M et al: Polar body array CGH for prediction of the status of the corresponding oocyte. Part II: technical aspects. Hum Reprod 2011; 26: 3181–3185.
Gutierrez-Mateo C, Colls P, Sanchez-Garcia J et al: Validation of microarray comparative genomic hybridization for comprehensive chromosome analysis of embryos. Fertil Steril 2011; 95: 953–958.
Benkhalifa M, Kasakyan S, Clement P et al: Array comparative genomic hybridization profiling of first-trimester spontaneous abortions that fail to grow in vitro. Prenat Diagn 2005; 25: 894–900.
Micale M, Insko J, Ebrahim SA, Adeyinka A, Runke C, Van Dyke DL : Double trisomy revisited – a multicenter experience. Prenat Diagn 2010; 30: 173–176.
Allen EG, Freeman SB, Druschel C et al: Maternal age and risk for trisomy 21 assessed by the origin of chromosome nondisjunction: a report from the Atlanta and National Down Syndrome Projects. Hum Genet 2009; 125: 41–52.
Golzio C, Guirchoun J, Ozilou C et al: Cytogenetic and histological features of a human embryo with homogeneous chromosome 8 trisomy. Prenat Diagn 2006; 26: 1201–1205.
Hassold T, Hunt P : Maternal age and chromosomally abnormal pregnancies: what we know and what we wish we knew. Curr Opin Pediatr 2009; 21: 703–708.
Garcia-Cruz R, Brieno MA, Roig I et al: Dynamics of cohesin proteins REC8, STAG3, SMC1 beta and SMC3 are consistent with a role in sister chromatid cohesion during meiosis in human oocytes. Hum Reprod 2010; 25: 2316–2327.
Jin F, Hamada M, Malureanu L et al: Cdc20 is critical for meiosis I and fertility of female mice. PLoS Genet 2010; 6.
Hulten MA, Patel S, Jonasson J et al: On the origin of the maternal age effect in trisomy 21 Down syndrome: the Oocyte Mosaicism Selection model. Reproduction 2010; 139: 1–9.
Conn CM, Cozzi J, Harper JC et al: Preimplantation genetic diagnosis for couples at high risk of Down syndrome pregnancy owing to parental translocation or mosaicism. J Med Genet 1999; 36: 45–50.
Roberts R, Iatropoulou A, Ciantar D et al: Follicle-stimulating hormone affects metaphase I chromosome alignment and increases aneuploidy in mouse oocytes matured in vitro. Biol Reprod 2005; 72: 107–118.
Bean CJ, Hassold TJ, Judis L et al: Fertilization in vitro increases non-disjunction during early cleavage divisions in a mouse model system. Hum Reprod 2002; 17: 2362–2367.
Acknowledgements
We acknowledge the expert support by Dr Maria Köster, Professor Katrin van der Ven, Professor Hans van der Ven (University of Bonn).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Professor Alan Handyside is a part time employee of Bluegnome Ltd. The other authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Handyside, A., Montag, M., Magli, M. et al. Multiple meiotic errors caused by predivision of chromatids in women of advanced maternal age undergoing in vitro fertilisation. Eur J Hum Genet 20, 742–747 (2012). https://doi.org/10.1038/ejhg.2011.272
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2011.272
Keywords
This article is cited by
-
MiR-425-5p suppression of Crebzf regulates oocyte aging via chromatin modification
GeroScience (2023)
-
New insights regarding origin of monosomy occurrence in early developing embryos as demonstrated in preimplantation genetic testing
Molecular Cytogenetics (2022)
-
Prenatal diagnosis of 7 cases with uniparental disomy by utilization of single nucleotide polymorphism array
Molecular Cytogenetics (2021)
-
Quercetin promotes in vitro maturation of oocytes from humans and aged mice
Cell Death & Disease (2020)
-
Natürliche Fertilität und Alter
Gynäkologische Endokrinologie (2020)