Abstract
Quinolones are used extensively to treat Shigella flexneri infection, and plasmid-mediated quinolone resistance (PMQR) determinants were recently reported. A total of 26 S. flexneri isolates collected from Anhui province, China, in 2005 were screened for PMQR determinants, bla gene, gyrA and parC genes, by PCR and sequencing. PMQR determinants were present in 53.8% (14 of 26) of isolates and qnrA1, qnrS1, qnrS2, aac(6′)-Ib-cr and qepA were present in 30.8, 11.5, 3.8, 19.2 and 11.5% of those isolates, respectively. All PMQR determinants' positive isolates exhibiting 8 different genetic clones had mutations in gyrA and parC genes, and 11 carried bla genes, including 7 blaCTX−M with resistance to cefotaxime. These isolates were resistant to nalidixic acid and 57.1% (8 of 14) of them were resistant to ciprofloxacin and levofloxacin. Our data show that there was a high prevalence of PMQR determinants in S. flexneri isolates from China.
Similar content being viewed by others
Introduction
Shigella flexneri is a causative agent of bacillary dysentery, especially in China, and quinolones are among the most important antibacterial agents used extensively for treating this infection.1, 2, 3 Recently, quinolone resistance has been increasing markedly in these isolates.2, 3 The common mechanisms for resistance to quinolone in pathogenic bacteria including S. flexneri are mutations in the chromosomal genes of DNA gyrase and DNA topoisomerase IV, the targets of quinolones.4 Efflux pumps and alteration in outer membrane proteins are also among the effects of chromosomal quinolone resistance.4
However, since 1998,5 three kinds of plasmid-mediated quinolone resistance (PMQR) determinants have been described: Qnr, AAC(6′)-Ib-cr and QepA.6, 7, 8 Several families of qnr (qnrA, qnrB, qnrS, qnrC, qnrD) have been reported and each family has different subtypes.6, 9 qnr encoded a protein of the pentapeptide family, protecting DNA gyrase directly from quinolone inhibition.10 AAC(6′)-Ib-cr is a variant of aminoglycoside acetyltransferase AAC(6′)-Ib with enzymatic modification of ciprofloxacin or norfloxacin.7 qepA encoded an efflux pump belonging to the major facilitator subfamily and quinolones were the specific substrates of QepA.8
To investigate the prevalence of PMQR determinants in S. flexneri isolates from China, we screened the prevalence of qnr, qepA and aac(6′)-Ib-cr genes by PCR and sequencing of positive PCR products in S. flexneri clinical isolates collected from Anhui province, China in 2005.
Materials and methods
Bacteria
A total of 26 consecutive clinical isolates of S. flexneri were collected from nine hospitals in Anhui province, China in 2005: Escherichia coli ATCC (American Type Culture Collection, Manassas, VA, USA) 25922, PMQR determinants' positive control strains and bla-positive control strains.
Antimicrobial agents
The following antimicrobial agents were used: ciprofloxacin, levofloxacin E-test strip (AB Biodisk, Solna, Sweden), cefotaxime, ceftazidime (National Institute for Control of Pharmaceutical and Biological Products, Beijing, China) and nalidixic acid (Sigma, St Louis, MO, USA).
Screening of qnr, qepA, aac(6′)-Ib-cr, gyrA, gyrB, parC, parE and bla genes
Amplification of qnr, qepA and aac(6′)-Ib-cr was performed using the methods described previously;8, 9, 11, 12 different primers (shown in Table 1) were used for the amplification of qnrB and qnrA subtypes in this study. Amplification of bla genes (β-lactamase genes) was also performed with the methods described previously for blaTEM, bla SHV and blaCTX−M.13, 14, 15 The mutations in the quinolone resistance-determining regions of the gyrA, gyrB, parC and parE genes were amplified by PCR.16 All purified PCR products were directly sequenced using an ABI3100 genetic analyzer and continued using primers walking on both DNA strands. For sequence comparison, the NCBI BLAST program and facilities of the TIGR Comprehensive Microbial Resource (http://www.tigr.org, National Center for Biotechnology Information, Bethesda, MD, USA) were used.
Antimicrobial susceptibility tests
Minimum inhibitory concentrations (MICs) of ciprofloxacin and levofloxacin were determined using E-tests. Susceptibility testing for cefotaxime, ceftazidime and nalidixic acid was performed by the agar dilution method according to the guidelines recommended by the Clinical and Laboratory Standards Institute.17 E. coli ATCC 25922 was used as the quality control strain.
Molecular typing of bacterial DNA
To elucidate the clonality of PMQR determinants' positive isolates, genomic DNA of each PMQR determinants' positive strain was analyzed by pulsed-field gel electrophoresis (PFGE) in a CHEF-MAPPER apparatus (Bio-Rad Laboratories, Hercules, CA, USA) after digestion with XbaI.15, 18 Genetic clones were identified according to criteria established by Tenover et al.18
Results and Discussion
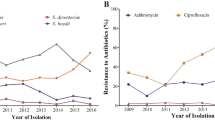
PMQR determinants were present in 53.8% (14 of 26) of isolates with qnr, aac(6′)-Ib-cr and qepA detected alone or in combination (Table 2). qnrA1, qnrS1, qnrS2, aac(6′)-Ib-cr and qepA were present in 30.8% (8 of 26), 11.5% (3 of 26), 3.8% (1 of 26), 19.2% (5 of 26) and 11.5% (3 of 26) of those isolates, respectively. Five isolates carried two or three types of PMQR determinants. In 14 PMQR determinants' positive isolates, all had the mutations in gyrA (S83L and/or D87N) and parC (S80I) genes, and the ciprofloxacin MICs of these strains were relatively high, 0.25 to >32 μg ml−1; of these, nine strains had MICs 2 to >32 μg ml−1. A total of 11 carried bla genes (most of the strains carried more than one type), including 7 blaCTX−M (6 blaCTX−M−14 and 1 blaCTX−M−15), 9 blaTEM−1 and 1 blaSHV−1, in all 14 PMAR-positive strains simultaneously. All PMQR determinants' positive isolates were resistant to nalidixic acid and 57.1% (8 of 14) of them were resistant to ciprofloxacin and/or levofloxacin. Seven blaCTX−M-positive strains were resistant to cefotaxime, only two of seven were resistant to ceftazidime according to CLSI standards.17 Results of PFGE show that 14 PMQR determinants' positive isolates exhibited eight different genetic clones by Tenover et al.18 criteria.
In 2005, qnrS1 was first described in S. flexneri isolates from Japan.19 The qnrS1 gene has ∼60 and ∼50% homology to the gene of qnrA and qnrB, respectively. There are 17 amino-acid substitutions between QnrS1 and QnrS2, but only one amino-acid substitution between QnrS1 and QnrS3.6 qnrC and qnrD are novel qnr-like genes and encoded 221 amino-acid and 214 amino-acid pentapeptide repeat proteins, respectively, with similarities to qnrA, qnrB and qnrS.9, 11 aac(6′)-Ib-cr and qepA genes are the second and third most recently detected PMQR determinants differing from the qnr family and are being increasingly identified worldwide among various clinical isolates of Enterobacteriaceae.7, 8, 12, 20 To our knowledge, this study is the first report of qnrA1, qnrS2, aac(6′)-Ib-cr and qepA genes in S. flexneri (the nucleotide sequence reported herein has been registered with the EMBL database under accession nos. FJ177519, FJ177521, FJ949571, FJ842530, respectively). The emergence of PMQR determinants in S. flexneri may affect the use of quinolones in the treatment of dysentery, and this study provides some new and useful information.
Most of the PMQR determinants provide only low-level resistance to fluoroquinolones, but can supplement to other quinolone-resistance mechanisms, such as the selection of chromosomal mutations in the presence of a quinolone.4, 5, 7, 8 PMAR determinants confer low-level resistance to quinolones with ciprofloxacin MICs 0.1–1.0 μg ml−1 in E. coli J53. For high-level ciprofloxacin- or levofloxacin-resistant strains, additional mechanisms such as target mutations and/or active efflux pump frequently coexist with PMQR determinants.7 In this study, mutations in gyrA and parC genes were identified in all PMQR determinants' positive isolates with resistance to nalidixic acid, and more than half of which were resistant to ciprofloxacin by CLSI criteria.17 S83L, D87Y mutations in gyrA and S80I in parC are the most common point mutations identified to cause quinolone resistance in several previous studies,4, 16, 21 and mutants with a single gyrA mutation had ciprofloxacin of MIC 0.32 μg ml−1.21 PMQR determinants are usually associated with additional resistant determinants and have often been reported to be coassociated with ESBL (extended-spectrum β-lactamase) genes and plasmid-mediated AmpC genes.4 More than half of the PMQR determinants associated with bla genes in this study, especially blaCTX−M, which conferred coresistance to the third generation cephalosporins. CTX-M-ESBLs (especially CTX-M-14) prefer hydrolyzing cefotaxime than ceftazidime and are the main type of ESBLs in Enterobacteriaceae isolates, including Shigella spp., in China.3, 13, 14
Results of PFGE show clonal spread in a few of these strains, which poses a serious threat to clinic. Some isolates (05ShC15/05ShC42 and 05FuY2/05FuY17) carry the same resistance genes, but the strains are in different PFGE types. It is possible that they were invaded by the same plasmid, which we will study in the future. Strains 05ShC24 and 05ShC25 are both in PFGE group I, and both carry aac(6′)-Ib-cr and blaTEM-1, but are different in CIP and LVFX. There may be other forms of chromosomal resistance, such as alteration in the outer membrane proteins.4 Of three PFGE H types, two strains had two mutations at both 83 and 87 positions in gyrA, whereas one strain only had a mutation at 83. The explanation might be that PFGE detects the molecular typing of bacterial DNA only by big DNA fragment mappers18 and those strains isolated from different hospitals.
Our data show that PMQR determinants were prevalent and qnrA1, qnrS2, aac(6′)-Ib-cr and qepA genes were first reported in S. flexneri from China. Most of those isolates are coresistant and clone spread exists in a few of them. It is necessary to monitor these isolates closely.
References
Turner, S. A., Luck, S. N., Sakellaris, H., Rajakumar, K. & Adler, B. Role of attP in integrase-mediated integration of the Shigella resistance locus pathogenicity island of Shigella flexneri. Antimicrob. Agents Chemother. 48, 1028–1031 (2004).
Vasilev, V., Japheth, R., Yishai, R. & Andorn, N. Antimicrobial resistance of Shigella flexneri serotypes in Israel during a period of three years: 2000–2002. Epidemiol. Infect. 132, 1049–1054 (2004).
Xiong, Z., Li, T., Li, H., Xu, Y. & Li, J. The genotype of extended-spectrum β-Iactamases in Shigella flexneri isolates. Chin. J. Infect. Dis. 24, 296–300 (2006).
Jacoby, G. A. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 15, S120–S126 (2005).
Martinez-Martinez, L., Pascual, A. & Jacopy, G. A. Quinolone resistance from a transferable plasmid. Lancet 351, 197–199 (1998).
Jacoby, G. A. et al. qnr gene nomenclature. Antimicrob. Agents Chemother. 52, 2297–2299 (2008).
Robicsek, A. et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12, 83–88 (2006).
Yamane, K. et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51, 3354–3360 (2007).
Wang, M. et al. New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 53, 1892–1897 (2009).
Tran, J.H., Jacoby, G.A. & Hooper, D. C. Interaction of the plasmid-encoded Quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob. Agents Chemother. 49, 118–125 (2005).
Cavaco, L. M., Hasman, H., Xia, S. & Aarestrup, F. M. qnrD, a novel gene conferring transferable quinolone resistance in Salmonella enterica serovar Kentucky and Bovismorbificans strains of human origin. Antimicrob. Agents Chemother. 53, 603–608 (2009).
Xiong, Z. et al. Investigation of qnr and aac(6′)-Ib-cr in Enterobacter cloacae isolates from Anhui Province, China. Diagn. Microbiol. Infect. Dis. 62, 457–459 (2008).
Xiong, Z. et al. A Klebsiella pneumoniae producing three kinds of class A β-lactamases encoded by one single plasmid isolated from a patient in Huashan Hospital, Shanghai, China. Int. J. Antimicrob. Agents 23, 262–267 (2004).
Xiong, Z., Li, T ., Xu, Y . & Li, J. Detection of CTX-M-14 extended-spectrum β-lactamase in Shigella sonnei isolates from China. J. Infect. 55, e125–e128 (2007).
Xiong, Z. et al. Investigation of extended-spectrum β-lactamase in Klebsiellae pneumoniae and Escherichia coli from China. Diagn. Microbiol. Infect. Dis. 44, 195–200 (2002).
Dutta, S. et al. Alteration in the GyrA subunit of DNA gyrase and the ParC subunit of topoisomerase IV in quinolone-resistant Shigella dysenteriae serotype I clinical isolates from Kolkata, India. Antimicrob. Agents Chemother. 49, 1660–1661 (2005).
CLSI. Performance Standards for Antimicrobial Susceptibility Testing: 17th Informational Supplement. M100–S17 (Clinical and Laboratory Standards Institute, Wayne, PA: Clinical and Laboratory Standards Institute, 2007).
Tenover, F. C. et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33, 2233–2239 (1995).
Hata, M. et al. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 49, 801–803 (2005).
Ma, J. et al. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac(6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicob. Agents Chemother. 53, 519–524 (2009).
Hopoper, D. C., Wolfson, J. S., Ng, E. Y. & Swartz, M. N. Mechanisms of action of and resistance to ciprofloxacin. Am. J. Med. 82, 12–20 (1987).
Acknowledgements
This study was supported in part by grants from the Key Technology Program of the Ministry of Education (209059), Outstanding Youth fund of Anhui province (08040106815) and from the Department of health, human research and social security of Anhui province, China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xiong, Z., Li, J., Li, T. et al. Prevalence of plasmid-mediated quinolone-resistance determinants in Shigella flexneri isolates from Anhui Province, China. J Antibiot 63, 187–189 (2010). https://doi.org/10.1038/ja.2010.16
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2010.16
Keywords
This article is cited by
-
Newest data on fluoroquinolone resistance mechanism of Shigella flexneri isolates in Jiangsu Province of China
Antimicrobial Resistance & Infection Control (2017)