Abstract
Spinocerebellar ataxia type 15 (SCA15) is an autosomal dominant neurodegenerative disorder clinically characterized by late-onset, slowly progressive pure cerebellar ataxia. This disease is caused by a heterozygous deletion of the inositol 1, 4, 5-triphosphate receptor type 1 (ITPR1) gene, suggesting that haploinsufficiency of the receptor function is the plausible disease mechanism. To clarify the prevalence of SCA15 in Japan, we designed four sets of probes and primers in different regions of ITPR1 and performed TaqMan PCR assay to search for gene deletions in 226 index SCA patients excluded for repeat expansion disorders. Deletion was found in only one patient, in whom gait ataxia started at 51 years of age and progressed to show cerebellar ataxia. This study demonstrates a simple but efficient method for screening ITPR1 deletion. We also conclude that ITPR1 gene deletions are much rare in Japan than in Europe, comprising only 0.3% in all SCAs in Japan.
Similar content being viewed by others
Introduction
Spinocerebellar ataxia (SCA) is a group of autosomal dominant neurodegenerative disorders clinically showing adult-onset progressive cerebellar ataxia as a cardinal feature.1 Extracerebellar signs, such as ophthalmoparesis, pyramidal tract, extrapyramidal signs or peripheral neuropathy may be variably seen. To date, 30 SCA gene loci have been identified.1, 2, 3 Among these, the expansion of tri-nucleotide (CAG) repeat accounts for the most common type of mutation and is the cause of SCA1, SCA2, SCA3/Machado–Joseph disease (MJD), SCA6, SCA7, SCA17 and dentatorubral-pallidoluysian atrophy (DRPLA). These disorders are called polyglutamine diseases, because the CAG repeat resides in the coding regions and is translated into polyglutamine tract. The second common mutation is the noncoding repeat expansion responsible for SCA8, SCA10, SCA12, SCA31 and probably also for recently identified SCA36.3 In contrast to these repeat expansion disorders, a few families are discovered to be caused by conventional, static mutations (for example, missense, frameshift or deletion) in functionally important genes. These include SCA5, SCA11, SCA13, SCA14, SCA15, SCA20, SCA23, SCA27, SCA28 and SCA35.2 Mutations in the remaining SCAs (SCA4, SCA18, SCA19, SCA21, SCA22, SCA25, SCA26, and SCA30) have not been discovered, and linkage assignment has not been established in still a significant number of families.
Identification of mutations is an essential prerequisite for exploring pathogenic mechanisms of these intractable diseases. On the other hand, the increase in the number of causative genes makes molecular test a laborious task from a standpoint of genetic diagnosis. Besides, information of clinical features is not always useful for prioritizing which genes to check, because clinical phenotypes of SCAs often overlap. Therefore, it is important to develop a simple and efficient method to screen mutations for SCA, particularly for those with static mutations.
Towards establishing efficient way to screen SCA mutations, we decided to examine SCA15 first. This disease is caused by mutations of the 5′ lesion of the inositol 1, 4, 5-triphosphate receptor type 1 (ITPR1) gene. Although one missense mutation has been reported,4 the vast majority of mutation is heterozygous deletions.5 Thus, we focused on screening deletions of ITPR1 and attempted to clarify the frequency of deletion in our cohort. Single-nucleotide mutations causing the rest of SCAs are going to be studied separately, as well as other rare repeat diseases such as SCA10.
Materials and methods
Patient enrollment
We collected peripheral blood samples from a total of 484 patients, all from unrelated families for purposes of genetic diagnosis at the Department of Neurology and Neurological Science, Tokyo Medical and Dental University, between 1 January 2004 and 31 October 2011. From each patient, blood was drawn after obtaining informed consent. Genomic DNA was extracted from white blood cells by standard phenol-chloroform extraction. The study conformed to the Tenets of the Declaration of Helsinki and was ethically approved by the institutional review board of Tokyo Medical and Dental University, Tokyo, Japan.
The 484 cases include not only familial cases but also sporadic cases, because incomplete penetrance in SCA may sometimes appear as sporadic ataxia, or sometimes a detailed family history may not be disclosed because of multiple reasons including private situations. Among these 484 cases, 258 patients were found to be SCA1, SCA2, MJD, SCA6, SCA31 or DRPLA. The remaining 226 patients were then tested for ITPR1 deletion. The breakdown of the 226 patients is as follows: 44 index cases with definite family history of autosomal dominant inheritance, 30 index cases with a certain family history with undetermined pattern of inheritance and 152 cases who did not have apparent familial history of similar disease (‘sporadic’). Patients with a diagnosis of multiple system atrophy at the time of genetic testing were not included in the 484 cases of this study.
Detection of ITPR1 copy number alterations
Quantitative PCR of genomic DNA was carried out by the TaqMan PCR assay, which was performed using a LightCycler480 system II (Roche Molecular Diagnostics, Basel, Switzerland) in a final volume of 20 μl reaction mixture containing 10 μl of LightCycler 480 Probes Master (Roche Molecular Diagnostics), 900 nM of each forward and reverse primer, 250 nM TaqMan probe and 30 ng of genomic DNA. Cycling conditions consisted of incubation at 95 °C for 10 min, and 40 cycles of 95 °C for 10 s, 60 °C for 30 s and 72 °C for 1 s. In each run, the LightCycler 480 software plotted the fluorescence intensity against the number of cycles and provided the crossing point (Cp) value.
Four sets of primer pairs and corresponding probes (sets 1–4) were designed in the ITPR1 genomic region (Figure 1). To securely detect various deletions, four sets were spaced so that at least two sets can detect 13 previously found deletions. Set 1, 2, 3 and 4 were designed at ITPR1 exon 2, intron 4, exon 6 and intron 9, respectively. The β-globin gene (HBB) was amplified as an endogenous reference. Details of nucleotide sequences of primers and probes are shown in Table 1. In addition, a genomic DNA sample of a previously described SCA15 subject4 was simultaneously tested as a positive control. This individual had been confirmed to harbour a total deletion of ITPR1 in one allele by two of us (OO and MN). The gene dosage strategy was based on the relative amplification of ITPR1 against HBB. The gene copy number was given by the formula 2−ΔΔCp; ΔCp=Cp(ITPR1)−Cp(HBB), ΔΔCp=ΔCp(sample)−ΔCp(negative control). The gene copy number between 0.4 and 0.6 was considered as heterozygous deletion, whereas the number between 0.9 and 1.1 was regarded normal.
An analysis of ITPR1 deletions in the present cohort of SCA. Genetic structures of SUMF1 and ITPR1 are shown on top. Vertical arrows with dotted lines show the recognition loci of the four sets of TaqMan probe and primers (Table 1) designed in this study. Horizontal line designates the region of deletion in each SCA15 family. Two thick lines indicate Japanese SCA15 families with ITPR1 deletion.
Calculation of ITPR1 deletion frequency in our cohort
The frequency of ITPR1 deletion among our cohort was finally expressed in two patterns: one was the frequency among ‘patients with family history’ (that is, patients with definite autosomal dominant inheritance or those with a certain family history) and the other was among ‘all patients’ (n=484) including seemingly sporadic cases.
Results
We first confirmed that the present sets of TaqMan primers and probes faithfully detect heterozygous deletion in ITPR1 (Figure 2). We then screened ITPR1 gene dosage in our cohort of 226 patients. None of these patients harboured deletions, except for one patient (TMDU-2010#055) who had a heterozygous deletion detected by all the four probe sets (Figures 2 and 3).
The present assay faithfully detects ITPR1 deletion. The present four sets of TaqMan probe detected ITPR1 deletion in a patient already described by Hara et al.,4 but not in negative control without ataxia. We also found again a heterozygous ITPR1 gene deletion in one (TMDU-2010#055) out of the entire cohort by all the sets of TaqMan primers and probes. Vertical bar indicates the gene copy number.
Screening for ITPR1 deletion with set 1. Half gene dosage is seen in one patient (TMDU-2010#055) as well as in positive control (SCA15 patient)4. Vertical axis indicates gene copy number.
Clinical features of the patient with heterozygous ITPR1 deletion (TMDU-2010#055) will be described in detail separately. Briefly, this patient first developed gait disturbance at the age of 51 years. Then the symptom gradually worsened. The patient's neurological features, examined by two of us (YI and RK), could be summarized as cerebellar ataxia, truncal tremor and increased deep tendon reflex in legs. Three out of five siblings also had similar ataxic symptoms. Though detailed clinical information of their parents was unavailable, neither parent showed apparent ataxic signs. It should be noted that this family did not meet criteria of definite autosomal-dominant ataxia.
As far as we are aware, only two families had been previously reported4, 6 as SCA15 with ITPR1 deletion from Japan. To compare the range of ITPR1 deletion between our case and the two families, we designed new sets of TaqMan probes and primers (Supplementary Table 1), which located near borders of the two reported deletions as follows: SUMF1 exon 2, SUMF1 exon 1, ITPR1 exon48 and ITPR1 exon59, and checked them. Our case preserved all these genes including ITPR1 exon 48, which is missing in both of the two families. Therefore, the deletion in our case is different from those in the two families.
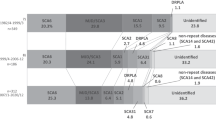
Our finding also demonstrates that SCA15 due to ITPR1 deletion is extremely rare in Japan, comprising approximately 0.2% of ‘all patients’ (when analysed including seemingly sporadic cases) and 0.3% of ‘patients with family history’ (Figure 4). This also indicates that SCA15 is much rare in Japan than in Europe.
Mutation frequencies in our cohort collected from 1 January 2004 to 31 October 2011. (a) SCA15 with ITPR1 deletion accounts for 0.2% of all the 484 patients including seemingly sporadic cases (for definition, please see Materials and Methods). (b) SCA15 with ITPR1 deletion accounts for 0.3% of patients with family histories of ataxia (332 cases). The disease frequencies shown in these figures, particularly those of SCA6 and 31, may be influenced by collecting bias.
Discussions
As summarized in Figure 1, 13 different ITPR1 gene deletions have been previously described.4, 6, 7, 8, 9, 10, 11 As shown in the present study, we confirmed that the present probe sets could detect ITPR1 deletion, and we further identified a new family with such mutation. It should be reminded that the present TaqMan system could overlook small deletions in ITPR1, but such small deletion is thought to be extremely rare from the following two reasons. One is that Hara et al.4 checked 54 SCA families for small deletions in ITPR1 with array comparative genomic hybridization but found no deletion. The other is that previous studies using multiplex ligation-dependent prove amplification assay, a comprehensive technique detecting small deletion in any exons, did not find such small deletions in their cohort.9, 10 From these points, we consider that the present screening system using four sets of primers and probes is a feasible and efficient technique to screen heterozygous ITPR1 deletions in a large sample set.
The frequency of ITPR1 deletion was very rare (0.3% of familial ataxia) in our cohort of patients. This confirms the previous finding made by Hara et al.4 that this disease is rare in Japan. When compared with the frequencies of ITPR1 deletion in Caucasian populations, the value we found is very low: reported frequencies of the deletion were 1.8% of autosomal dominant SCAs in Germany,9 3.2% of all SCAs including sporadic cases in Australia10 and 1.8% of autosomal dominant SCAs in France.12 In Italy, Di Gregorio et al.8 reported two SCA15 families: one out of 60 index cases from those having family history of ataxia and another one with a definite autosomal-dominant inheritance. They did not describe the frequency, but 3.3% (2/61) is similar to other reports on the frequency of SCA15 in Caucasian populations. These reports and the present did not examine missense mutations. As has been already described in Introduction, only one SCA15 family with a missense mutation in ITPR1 has been previously reported.4 For establishing a true frequency of SCA15, we need to check the missense mutation, although the vast majority of mutation in SCA15 is heterozygous deletions.5
Finally, the remaining families in our cohort of SCAs are excluded for SCA1, SCA2, MJD, SCA6, SCA15, SCA31 and DRPLA. A majority of these families showed pure cerebellar ataxia with variable ages of onset. It would be prudent to examine other SCAs with relatively pure cerebellar syndrome, such as SCA5, 8, 10, 14 and 35. However, many of the remaining SCAs are caused by static mutations, making it a laborious task to accomplish for more than 200 samples. Because their clinical features lack any of the characteristic features of rare SCAs, such as anticipation and pyramidal tract signs in SCA5, epilepsy in SCA10, myoclonus or tremor in SCA14, it is not surprising even if most of our families are proven to be none of the remaining SCAs with known mutations. To make genetic diagnosis of SCA more feasible and cost-effective, it appears important to delineate clinical features of ‘unknown’ SCAs (that is, SCAs without known mutations) to help prioritizing which SCAs should be tested first. In parallel, challenges to identify novel gene mutations in these ‘unknown’ SCAs are warranted.
References
Durr, A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol. 9, 885–894 (2010).
Wang, J. L., Yang, X., Xia, K., Hu, Z. M., Weng, L., Jin, X. et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain. 133, 3510–3518 (2010).
Kobayashi, H., Abe, K., Matsuura, T., Ikeda, Y., Hitomi, T., Akechi, Y. et al. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am. J. Hum. Genet. 89, 121–130 (2011).
Hara, K., Shiga, A., Nozaki, H., Mitsui, J., Takahashi, Y., Ishiguro, H. et al. Total deletion and a missense mutation of ITPR1 in Japanese SCA15 families. Neurology. 71, 547–551 (2008).
van de Leemput, J., Wavrant-De Vrieze, F., Rafferty, I., Bras, J. M., Giunti, P., Fisher, E. M. et al. Sequencing analysis of the ITPR1 gene in a pure autosomal dominant spinocerebellar ataxia series. Mov. Disord. 25, 763–765 (2010).
Iwaki, A., Kawano, Y., Miura, S., Shibata, H., Matsuse, D., Li, W. et al. Heterozygous deletion of ITPR1, but not SUMF1, in spinocerebellar ataxia type 16. J. Med. Genet. 45, 32–35 (2008).
van de Leemput, J., Chandran, J., Knight, M. A., Holtzclaw, L. A., Scholz, S., Cookson, M. R. et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 3, 1076–1082 (2007).
Di Gregorio, E., Orsi, L., Godani, M., Vaula, G., Jensen, S., Salmon, E. et al. Two Italian families with ITPR1 gene deletion presenting a broader phenotype of SCA15. Cerebellum. 9, 115–123 (2009).
Synofzik, M., Beetz, C., Bauer, C., Bonin, M., Sanchez-Ferrero, E., Schmitz-Hubsch, T. et al. Spinocerebellar ataxia type 15: diagnostic assessment, frequency, and phenotypic features. J. Med. Genet. 48, 407–412 (2011).
Ganesamoorthy, D., Bruno, D. L., Schoumans, J., Storey, E., Delatycki, M. B., Zhu, D. et al. Development of a multiplex ligation-dependent probe amplification assay for diagnosis and estimation of the frequency of spinocerebellar ataxia type 15. Clin. Chem. 55, 1415–1418 (2009).
Novak, M. J., Sweeney, M. G., Li, A., Treacy, C., Chandrashekar, H. S., Giunti, P. et al. An ITPR1 gene deletion causes spinocerebellar ataxia 15/16: a genetic, clinical and radiological description. Mov. Disord. 25, 2176–2182 (2010).
Marelli, C., van de Leemput, J., Johnson, J. O., Tison, F., Thauvin-Robinet, C., Picard, F. et al. SCA15 due to large ITPR1 deletions in a cohort of 333 white families with dominant ataxia. Arch. Neurol. 68, 637–643 (2011).
Acknowledgements
We would like to thank Mrs Hitomi Matsuo for her technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Dr Ishikawa received research grants from the Kobayashi Magobei Research Foundation, Mitsubishi Zaidan Research Foundation, the Grants-in-Aid for Scientific Research (21591072 and 21249054), Japan Society for the Promotion of Science, Japan, Grant-in-Aid for Scientific Research on Innovative Areas (23129503) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Health and Labour Sciences Research Grants on Human Genome, Ministry of Health, Labour and Welfare, Japan. Dr Mizusawa received research grants from the Grants-in-Aid for Scientific Research (21249054), Japan Society for the Promotion of Science, Japan, the Strategic Research Program for Brain Sciences by the Ministry of Education, Culture, Sports, Science and Technology of Japan, the Core Research for Evolutional Science and Technology (CREST), Japan Science and Technology Agency (JST), Saitama, Japan, and the Health and Labour Sciences Research Grants Ataxic Diseases, Ministry of Health, Labour and Welfare, Japan.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Obayashi, M., Ishikawa, K., Izumi, Y. et al. Prevalence of inositol 1, 4, 5-triphosphate receptor type 1 gene deletion, the mutation for spinocerebellar ataxia type 15, in Japan screened by gene dosage. J Hum Genet 57, 202–206 (2012). https://doi.org/10.1038/jhg.2012.5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2012.5
Keywords
This article is cited by
-
A novel gain-of-function mutation in the ITPR1 suppressor domain causes spinocerebellar ataxia with altered Ca2+ signal patterns
Journal of Neurology (2017)
-
Sporadic infantile-onset spinocerebellar ataxia caused by missense mutations of the inositol 1,4,5-triphosphate receptor type 1 gene
Journal of Neurology (2015)