Abstract
The MMP2 gene has been implicated in the pathogenesis of endometriosis. We investigated the role and function of single-nucleotide polymorphisms (SNP) of MMP2 in relation to endometriosis. First a case–control study was conducted and 17 SNPs were examined in 211 patients and 344 controls. Regression analysis was used to evaluate the genetic effect. We used reporter assay to validate the functional consequences of the significant SNP. Two SNPs (rs243832 and rs7201) had P-values <0.05 and they are in strong linkage disequilibrium (D′=0.96 and r2=0.47). Further analysis showed that rs7201 but not rs246832 was an independent risk factor and the risk C allele of rs7201 had an odds ratio (OR) of 1.88 (P=0.004). SNP rs7201 is located at the 3′-untranslated region and is predicted to be within the microRNA-520g binding site. The reporter assay for rs7201 showed that the risk C allele had a higher expression level than the A allele (P=0.027). Using microRNA-520g mimic and inhibitor, the results indicated that the A allele but not the risk C allele can be regulated by microRNA-520g. The C allele of SNP rs7201 increases a risk for endometriosis because of out of regulation by microRNA-520g.
Similar content being viewed by others

Introduction
Endometriosis is a common gynecological disorder. The disease is characterized by the presence of endometrial glands and stroma outside their normal locations. Symptoms associated with endometriosis include dysmenorrhea, dyspareunia, non-cyclic pelvic pain and infertility. A growing body of evidence indicates that genetic factors influence susceptibility to the disease1, 2 and several genetic polymorphisms have been implicated for the risk of this disease.3 Several steps have been implicated for the development of endometriosis, which include retrograde menstruation, attachment of endometrial fragments to the epithelium of the peritoneum, invasion of the epithelium, establishment of a blood supply and generation of a suboptimum immune response that does not adequately clear the implants.4
Proteolytic activity of cell–surface proteins and extracellular matrix has a pivotal role in the biological process of menstruation and pathological process in the development of endometriosis. The process of cell migration or invasion is associated with the degradation and reconstruction of the surrounding microenvironment that consists of extracellular matrix components. The matrix metalloproteinases (MMPs) are a broad family of zinc-binding endopeptidases that are responsible for the degradation of extracellular matrix components in several organs/systems including female reproductive system.5 It was also shown that endometrial stromal cells secrete several MMPs, including MMP2.6, 7 The uterine endometrium from women with endometriosis expresses a higher level of MMP2.6, 7 Accumulating evidence suggests that MMP2 is involved in the formation of endometriosis.6, 7, 8, 9, 10
This study aimed to test for an association between polymorphisms of the MMP2 gene and to elucidate the underlying molecular mechanism of genetic variation in relation to endometriosis. We first used tagging single-nucleotide polymorphisms (tSNPs) plus a known functional SNP to evaluate systematically the association between the MMP2 gene and the disease. As genetic association studies are subject to a type I error, we further conducted a series of functional studies to support the statistical result from the association study. This study illustrated that microRNA-520g (miR-520g) and SNP rs7201 has a pivotal role in the regulation of MMP2 expression, which in turn confers risk for endometriosis.
Materials and methods
Genetic association study
Subjects
The study was approved by the Kaohsiung Medical University Hospital (KMUH) Institutional Review Board. All participants gave the informed consent. For the endometriosis cases, we only included the patients who underwent laparotomy or laparoscopy at KMUH and had pathological confirmation of the disease. All the cases were recruited from the Department of Obstetrics and Gynecology at the KMUH in Taiwan.
We compared the cases with two different control groups: sources 1 and 2. Source 1 included the normal controls who had regular menstruation, did not have any evidence of endometriosis by sonography and had no history of gynecological tumors upon enrollment. The controls from source 2 were recruited from communities. The details of the ascertainment scheme were described elsewhere.11 In brief, the subjects were the participants in community-based Pap smear screening projects.12, 13 An eligible control in this study did not have any of the following conditions: infertility, dysmenorrhea, hypermenorrhea, irregular menstruation, surgical history for any obstetrical diseases, previously diagnosed endometriosis or adenomyosis. Owing to insufficient DNA samples, some of the controls used in our previous studies11, 14 were not included in this study. All cases and controls are Chinese descents.
SNP selection and genotyping
One commonly studied functional polymorphism (−1306 C/T, rs243865)15 and 16 tSNPs selected from the HapMap Project (http://www.hapmap.org) were genotyped in this study. All tSNPs have the minor allele frequency ⩾10% in the Han Chinese population based on the HapMap database (www.hapmap.org). Some tSNPs indicated by the HapMap Project were replaced by our previously genotyped SNPs16 that were in strong linkage disequilibrium (r2⩾0.8) with the indicated tSNPs. Genotyping was carried out by using the TaqMan technology (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
A goodness-of-fit χ2 test was used to test for Hardy–Weinberg equilibrium (HWE) in the controls. We compared the mean of age, body mass index and parity between cases and controls. Because these three variables may also be associated with disease risk, they were included in the multivariate regression models. To account for multiple testing, we also performed family-wise permutation to obtain an empirical P-value. In this study, we used the minP permutation procedure to calculate the empirical P-values. This is a distribution-free statistic, which uses random label swapping to generate the null distribution in our population. The null distribution was obtained from the following steps: (1) swapping refractive errors randomly among all study subjects; (2) performing χ2 test (or Fisher’s test if a cell contains four or fewer samples) on the permuted data to obtain the minimal P-value of each run; and (3) repeating steps 1 and 2 to obtain 10 000 permutations. The empirical P-values for each genotype were calculated by the following formula: (R+1)/(N+1). R is the number of runs in which the minimal P-value is less than the observed; N is the number of permutations.
Odds ratio (OR) was calculated to assess the genetic effect. All quantitative data are presented as mean±standard deviation (s.d.). Statistical analyses were performed with the SPSS statistical software (version 12.0, SPSS Inc., Chicago, IL, USA). The Hap-Clustering program17 was used for haplotype analyses with adjustment for the three covariates.
Functional study
Reporter assay to assess the differential allelic effect on gene expression
A genomic DNA segment of 1004 bp that includes rs7201 was amplified using polymerase chain reaction (PCR) with the forward primer of 5′-AAAAAGCTTGGAAGCATCAAATCCGACTG-3′ and reverse primer of 5′-AAAGGATCCGCTTACTCTCCCTGAGGTTCTC-3′. The DNA segment was subcloned into the downstream (HindIII/BamHI sites) of the green fluorescent protein (GFP)-expressing vector, pEGFP-C1 (Clontech, Mountain View, CA, USA). Two reporter constructs were created. One construct carries the A allele and was designated as pEGFP-A, and the other carries C allele and was designated as pEGFP-C. The constructs were confirmed by DNA sequencing. Transient cotransfection was performed with 1 μg of each construct and 1 μg of pDsRed (Promega, Madison, WI, USA) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). The reporter assay was tested in the three endometrial cancer cell lines (RL95-2, HEC-1A and HEC1B) and one lung cancer cell line (H1299). pDsRed was used to calibrate the transfection efficiency. After 48 h, the GFP and DsRed signals were determined using a fluorometer (BioTek FLx800, Winooski, VT, USA). The GFP signal was detected as green (G) fluorescence and DsRed red (R) fluorescence, and we compared the ratio of G/R from each of the two reporter constructs. Each experiment was independently performed three times and each sample was studied in triplicates in each experiment. Statistical analysis was performed with the Mann–Whitney U-test with a two-side P-value ⩽5% as significance.
Immunohistochemical stain
We used the immunohistochemical (IHC) stain for pathological specimens from 24 cases with severe endometriosis (stages III and IV) according to the revised American Society for Reproductive Medicine (Re-ASRM). The technician who conducted the IHC experiment was blind to the clinical and genetic information. Each tissue specimen was routinely embedded in paraffin wax after 10% formalin fixation, and cut into 4-μm-thick section for MMP2 immunostaining. In brief, sections were deparaffinized and rehydrated. Endogenous peroxidase in the section was soaked in absolute methanol containing 3% hydrogen peroxide. After washing with Tris buffer solution, sections were incubated with the anti-MMP2 primary antibody. Sections were subsequently incubated with biotin-conjugated goat anti-mouse immunoglobulin G and avidin–biotin–peroxidase complex. Finally, the sections were incubated with 0.05% 3,3′-diaminobenzidine tetrahydrochloride (DAKO Cytomaion liquid DAB+Substrate chromogen System, DAKO Sytomation Inc., Carpinteria, CA, USA), followed by counterstained with Mayer’s hematoxylin and mounted with Histomount (Invitrogen). Antigen retrieval was carried out. Negative controls were obtained by replacing the primary antibody with non-immune serum, and the positive control sample was breast carcinoma.
The MMP2 IHC stain has species specificity, which means it only reacts with human MMP2 but does not react with mouse and rat.
The MMP2 IHC stain was evaluated by the semiquantitative score system: 0 indicating that the sample was not immunoreactive in the endometriotic cells; 1, 2, 3 and 4 indicating 1–25%, 26–50%, 51–75% and >75% immunoreactivity in the endometriotic cells, respectively. A positive stain was defined as a score ⩾3.
Measure endogenous miRNA in the cell lines
Three cell lines (H1299, RL95-2 and HEC-1A) were used to investigate miR-520g effect. First of all, we used the quantitative real-time PCR to measure the endogenous miR-520g levels. The microRNA Isolation Kit (mirVana; Applied Biosystems) was used to extract total RNA that also contains small RNAs. The TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) was used to convert microRNA to cDNA. Quantitative real-time PCR was performed on the ABI PRIZM 7500 sequence detection system (Applied Biosystems). The typical amplification program was carried out in a 20 μl reaction and real-time PCR results were analyzed using the ABI PRISM 7500 SDS software (Applied Biosystems). Each experiment was independently performed three times and each sample was studied in triplicates in each experiment. The Mann–Whitney U-test was used for statistical analysis with a two-side P-value ⩽5% as significance.
pEGFP-rs7201 reporter assay for differential knockdown effect by miR-520g
In all, 50 and 100 nM of miR-520g mimic (pre-miR miRNA precursor molecule; Ambion-Life Technologies, Carlsbad, CA, USA) were transfected along with 1 μg of reporter construct and 1 μg of pDsRed to the HEC-1A cells using the Lipofectamine 2000 reagent (Invitrogen). Similarly, the HEC-1A cells were transiently transfected with 50 and 100 nM of antagomiR-520g (Anti-miR miRNA Inhibitor; Ambion) and a negative control RNA (Ambion), respectively. miR-520g mimic is a synthetic RNA duplex and it acts as a mimic of naturally occurring miR-520g upon delivering into cells. AntagomiR-520g is a chemically engineered oligonucleotide to silence endogenous miR-520g. After 48 h, the GFP and DsRed signals were detected to determine the effects of miR-520g mimic and antagomiR-520g on the two reporter constructs. The results were analyzed using the Mann–Whitney U-test.
MMP2 gene is silenced by miR-520g
The effects of miR-520g mimic on MMP2 mRNA and protein expression were also evaluated in the primary endometrial stromal cells. The primary stromal cells were obtained from an endometriosis patient who carries the AA genotype at SNP rs7201. The stromal cells were grown in the Dulbecco’s modified Eagle’s medium/F12 medium supplemented with 10% fetal bovine serum. Cells between passages 3 and 6 were applied for the experiments. By using 5 μl siPort NeoFx (Ambion) reagent, concentrations of 30, 60 and 120 nM of miR-520g mimic were transfected into the stromal cells for 48 h. Quantification of MMP2 mRNA was conducted using High Capacity cDNA Reverse Transcription kit (Applied Biosystems) with random primers, 0.5 μg total RNA and the MultiScribe Reverse Transcripase according to the protocol. Gene-specific PCR primers and TaqMan probes for human MMP2 (ABI ID number: Hs01548731_m1) were from the Applied Biosystems. MMP2 mRNA expression was normalized to glyceraldehyde-3-phosphate dehydrogenase as an internal control. The protein extracts from the cultured stromal cells were determined by the western blot analysis. Equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A standard western blot analysis was conducted using MMP2 propeptide antibody (1:500 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA). β-Actin antibody (1:5000 dilution; Cell Signaling, Beverly, MA, USA) was used as a loading control.
Function prediction
Microcosm Targets version 5 database (http://www.ebi.ac.uk/enright-srv/microcosm, accessed in March 2010) was used to predict target genes of miR-520g. GeneOntology enrichment analysis was carried out using the MetaCore Pathways Analysis (GeneGo, Encinitas, CA, USA) by uploading the data set of predicted target genes to the MetaCore server. Target genes that were not expressed in the uterus were excluded from the MetaCore analysis. Enrichment analysis identified the most significant biological processes based on the uploaded target genes.
Results
Demographic data
This study included a total of 211 women with endometriosis, and 126 hospital-based and 218 community-based controls. According to Re-ASRM, 179 patients with endometriosis were in stages III and IV and 32 in stages I and II.
SNP and endometriosis
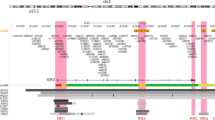
A total of 17 SNPs were selected for genotyping. Two SNPs are in exons (rs1053605 synonymous SNP, and rs243849 synonymous SNP), one SNP is in the 3′-untranslated region (UTR) (rs7201) and the rest SNPs are in the non-coding regions. All SNPs were in HWE except SNP rs9302671 (HW P-value=0.00035), and this SNP was not included for further genetic analyses. Based on χ2 or Fisher’s exact tests, only two SNPs had P-values <0.05: rs243832 (P=0.045 for the overall genetic effect and P=0.017 for the comparison between CC and GG genotypes) and rs7201 (P=0.014 for the overall genetic effect and the OR for AC was 1.88 with the P=0.004 for the comparison between AC and AA genotypes) (Table 1). The family-wise permutation was conducted to adjust for multiple testing and the empirical P-value for the AC genotype of SNP rs7201 was 0.047. Rs243832 and rs7201 SNPs are located in the same block with the D′=0.96 and r2=0.47 (Figure 1). When both SNPs were included in the regression model, only SNP rs7201 remained significant. We also used these two SNPs (rs243832 and rs7201) for the haplotype analysis and the result yielded a global P-value of 0.045.
In all, 17 MMP2 SNPs form four haplotype blocks. The r2 (%) between any two SNPs is displayed in each cell. The cells in dark gray indicate strong linkage disequilibrium and log of odds (LOD) of 2 or greater and in white indicate uninformative with LOD lower than 2. The bar on the top is the MMP2 gene structure and location of each SNP; kb indicates kilobase. SNPs rs1053605 and rs243849 are synonymous SNPs, SNP rs7201 is in the 3′-UTR and the res SNPs are not in the coding regions.
As MMP2 has been reported to be more related to severe endometriosis,10 we also analyzed 179 severe cases who were in Re-ASRM stage III or IV. The results for SNP rs243832 and rs7201 were less significant than those from all the cases (data not shown). Therefore, the association between MMP2 SNPs and endometriosis is not only restricted to the severe cases.
SNP rs7201 influences gene expression
Our statistical results suggest that SNP rs7201 was significantly associated with endometriosis. Because this SNP is located at 3′-UTR, we first conducted a reporter gene assay to test whether this SNP can influence gene expression levels. If so, we would further test whether this SNP could influence a microRNA binding. First, we compared GFP expression between two reporter constructs (pEGFP-A and pEGFP-C). GFP expression was found to be significantly higher for the pEGFP-C construct (G/R value was 2.30 for pEGFP-C and 1.62 for pEGFP-A, P=0.027 by Mann–Whitney U-test) transfected to the H1299 cell line. That is, the C allele yields a higher protein production than the A allele by 1.42-fold. Similarly, higher GFP expression for the pEGFP-C than pEGFP-A construct was shown in other three endometrial cancer cell lines: G/R ratio in the RL95-2 cell line was 1.99 for pEGFP-C and 1.78 for pEGFP-A; in the HEC-1A cell line was 1.89 for pEGFP-C and 1.73 for pEGFP-A; and in the HEC1B cell line was 1.90 for pEGFP-C and 1.33 for pEGFP-A.
MMP2 expression in different genotypes
We measured MMP2 expression in the endometrial samples from 24 patients with severe endometriosis. The number of subjects in the AA, AC and CC genotypes of SNP rs7201 was 16, 6 and 2 CC, respectively (Table 2). The P-value from the Mann–Whitney U-test was 0.027 for comparison between positive and negative IHC stains among three genotypes (Table 2). The CC subjects had higher mean IHC score and positive IHC stain, followed by the AC subjects and then AA subjects.
Endogenous miRNA-520g in the cell lines
Given that SNP rs7201 could influence gene expression, we searched for putative microRNAs that were predicted to bind to this locus. Using the bioinformatics database (http://mirnamap.mbc.nctu.edu.tw/ and http://www.microrna.org/microrna/home.do), both miR-520g and miR-520h were predicted to bind the 3′-UTR including the location of SNP rs7201 (Figure 2). In addition, both miR-520g and miR-520h are highly expressed in the placenta (http://mirnamap.mbc.nctu.edu.tw/). As miR-520g and miR-520h have a similar structure, we only investigated the miR-520g effect on the MMP2 gene. We measured the expression level of endogenous miR-520g in three cell lines (H1299, RL95-2 and HEC-1A) and primary stromal cells obtained from a single endometriosis patient. The endogenous miR-520g level was much higher (P<0.0001) in the stromal cells than three cancer cells (Supplementary Figure). Among the three cancer cell lines, the endogenous miR-520g levels were also different (P=0.007 for the comparison between RL95-2 and HEC-1A, P=0.04 between RL95-2 and H1299 and P=0.2 between HEC-1A and H1299).
SNP rs7201 influences gene expression via miRNA-520g
We further verified the effect of miR-520g on GFP expression by the overexpression and knockdown strategies. The experiments were conducted in three groups for each reporter construct: Group I—cotransfection of reporter constructs with negative control oligo (Invitrogen; Cat. No. AM17110), Group II—cotransfection of reporter constructs with miR-520g mimic and Group III—cotransfection of reporter constructs with the antagomiR-520g. After transfecting miR-520g mimic to the HEC-1A cells for 24 h, we detected a striking increase (P=0.0003) of intracellular miR-520g levels by more than 500-fold (Figure 3a). On the contrary, transfecting antagomiR-520g led to a significant decrease (P=0.0003) of endogenous miR-520g (Figure 3a). Similarly, miR-520g mimic and antagomiR-520g also caused comparable changes in the RL95-2 cells (data not shown). The above experiments indicated that mimic and antagomiR could significantly alter intracellular miR-520 levels.
(a) Transfecting miR-520g mimic and antagomiR-520g to the HEC-1A cells significantly changed the intracellular miR-520g levels (*P-value=0.0003). (b) The G/R ratio for the pEGFP-A construct was significantly reduced by miR-520g mimic in the HEC-1A cells. AntagomiR-520g increased the G/R ratio for the pEGFP-A construct. The G/R ratio for pEGFP-C was not influenced by miR-520g mimic. AntagomiR-520g barely increased the G/R ratio (P=0.3 for 50 vs 0 nM, P=0.04 for 100 vs 0 nM) for the pEGFP-C construct. *P<0.005. Each experiment was independently performed three times and each sample was studied in triplicates in each experiment.
The G/R ratio for the pEGFP-A construct was significantly suppressed by miR-520g mimic (P<0.005 for 50 vs 0 nM, P<0.005 for 100 vs 0 nM; Figure 3b). Using trend t-test, we found a significant dose-dependent manner (P<0.0001) for miR-520g silencing effect. Similarly, there was a significant increase (P<0.005 for 50 vs 0 nM, P<0.005 for 100 vs 0 nM; Figure 3b) of G/R ratio by antagomiR-520g for the pEGFP-A construct, and the increase also had a dose-dependent manner (P<0.0001). On the contrary, the G/R ratio was not or barely influenced by miR-520g mimic (P=0.4 for 50 vs 0 nM, P=0.4 for 100 vs 0 nM; Figure 3b) or antagomiR-520g (P=0.3 for 50 vs 0 nM, P=0.04 for 100 vs 0 nM; Figure 3b) for the pEGFP-C construct. Our results indicate that the risk C allele is less responsive to the miR-520s silencing effect because it causes imperfect complement to miR-520 seed region.
miR-520g directly represses MMP2 expression
To further confirm that the miR-520g can downregulate the MMP2 gene, we directly examined the influence of miR-520g on MMP2 expression in the primary endometrial stromal cells of the AA genotype at SNP rs7201. The result showed that both MMP2 mRNA and propeptide expressions were reduced in parallel with the concentrations of miR-520g mimic (Figure 4). Transfecting miR-520g mimic led to a decrease of MMP2 mRNA levels (compared with the baseline status, P<0.05 for 30 nM and all P-values <0.01 for 60, 90 and 120 nM; Figure 4a). The reduction of MMP2 mRNA expression also had a significant trend (P<0.0001) with the dose of miR-520g mimic. For MMP2 propeptide, miR-520 mimic reduced propeptide synthesis (compared with the baseline, P<0.05 for 30 nM and all P-values <0.01 for 60 or 120 nM; Figure 4b), which also showed a significant dose-dependent manner (P<0.0001). When 120 nM of miR-520g mimic was transfected, the propeptide production could be reduced by 45%.
(a) miR-520g mimic significantly downregulated MMP2 mRNA expression in the endometrial stromal cells, and the miR-520 effect also showed a dose-dependent manner. *P<0.05; **P<0.01 (b) miR-520g suppressed MMP2 propeptide expression in the endometrial stromal cells and the miR-520 effect on propeptide also had a dose-dependent manner (P<0.0001). *P<0.05; **P<0.01. Each experiment was independently performed three times and each sample was studied in triplicates in each experiment.
Discussion
In this study, we systematically investigated the relationship between 17 SNPs of the MMP2 gene and endometriosis. The previously known functional promoter polymorphism (rs243865) is not associated with endometriosis but the 3′-UTR SNP rs7201 is significant for endometriosis. Our IHC experiment also suggests differential expression of MMP2 in three different genotypes of SNP rs7201, and the risk C allele had a higher expression level. Both in silico and experimental studies indicate that rs7201 is located in the miR-520g binding site. We demonstrate that miR-520g mimic can downregulate the MMP2 expression. We also show that the risk C allele of SNP rs7201 acts as a loss-of-function mutation, because neither miR-520g mimic nor antagomiR-520g can influence the MMP2 expression in the presence of this allele. This loss-of-function mutation results in dysregulation of MMP2 expression and a higher risk for endometriosis. As dysregulation of MMP2 expression has been implicated as a pathogenesis of endometriosis,6, 8 our study provides an additional insight into the mechanism of endometriosis.
MMP2 is a highly expressed protein in endometrium, especially in ectopic endometrium.18 Women with endometriosis also have a higher level of MMP2 in the serum and peritoneal fluid than women without this disease.8 These observations are compatible with our finding in which the risk C allele causes a higher expression level and causes out of regulation by miR-520g. The MMP2 gene polymorphisms have been implicated in different diseases but most of previous studies only focused on functional promoter SNPs.19, 20, 21 Only a limited number of studies investigated endometriosis in relation to MMP2 polymorphisms.21, 22, 23 Analyzing the two functional promoter SNPs (rs2285053 also known as −735 C/T and rs243865 also known as −1306 C/T) in the Chinese patients with endometriosis, both Kang et al.22 and Zhao et al.23 did not find any significant association. Although Saare et al.21 reported that −735 C/T SNP was significant (P=0.02) for endometriosis, the significant result was primarily from 92 cases with Re-ASRM stages I and II but not from patients with Re-ASRM stages III and IV. On the other hand, we use the tagging SNP approach to investigate comprehensively the overall gene effect of MMP2. Although we do not disclose a relationship between the promoter polymorphism −1306 C/T and endometriosis, our study identifies another functional SNP rs7201 at the 3′-UTR conferring risk for endometriosis. Recently, the C allele of SNP rs7201 was also found to be an independent risk factor for stroke.24
SNP rs7201 is located in the complementary DNA region to the seed region of miR-520g. The MMP2 3′-UTR spans 1249 bp and only two validated SNPs are in the vicinity of SNP rs7201. The distance between rs7201 and rs17860019 is 101 base pairs (bp), and between rs7210 and rs17860021 is 140 bp. However, these two neighboring SNPs are too rare (rs17860019 is monomorphic in Asians; and the minor allele frequency of rs17860021 is about 1%) based on the NCBI dbSNP data set (131 version).
Identification the role of microRNAs in endometriosis opens a new avenue for biomarkers and therapeutic tools. Using global microRNA expression profiles in relation to endometriosis, recent studies did not find miR-520g or miR-520h to be differentially expressed between ectopic and eutopic endometrial tissues.25, 26, 27, 28 Notably, the expression of miR-520g and miR-520h is highly tissue specific. Both microRNAs are barely detectable in other tissues except for placenta (http://mirnamap.mbc.nctu.edu.tw/). Unfortunately, there are no available expression data for miR-520g or miR-520h in the uterus. The functional role of miR-520g is hardly mentioned in the literatures. From GeneOntology enrichment analysis, miR-520g target genes are enriched in cGMP-mediated signaling pathway, vasodilation and neural transmitters. The cGMP-mediated signaling pathway was reported to be associated with neointima thickness.29 Accordingly, miR-520g could have an important role in blood vessel function and/or development.
A SNP located at or near a microRNA binding site has been named as mirSNP, which represents a novel class of functional SNPs. Mishra et al.30 reported that a mirSNP located 14 bp downstream of the miR-24 binding site of the dihydrofolate reductase gene can affect dihydrofolate reductase expression. The mutant allele causes overexpression of dihydrofolate reductase protein, which is similar to this study. As dihydrofolate reductase is a target of methotrexate, patients with mutant allele at this mirSNP are resistant to methotrexate treatment. The application of mirSNP may lead to a different type of personalized medicine called microRNA pharmacogenomics.31 Recently, mirSNPs located in the WHSC1, SLC22A2332 and KRAS33 genes have been reported to influence the risk for endometriosis. As MMP2 has been implicated in several common diseases including cancer and cardiovascular disease,19, 20, 21, 34 using RNA interference for the subjects carrying the risk C allele of SNP rs7201 could be one of microRNA pharmacogenomic applications in the future.
There are several limitations in this study. Although adjusted (that is, empirical) P-value for the rs7201 AC genotype is 0.047 by the family-wise permutation test, readers still need to take the statistical results with caution because our sample size is still too moderate to avoid a type I error. However, functional studies in conjunction with SNP association studies can reduce such errors. The biosamples of the rare CC genotype at SNP rs7201 is hard to obtain. We only had two CC subjects for IHC stain and we do not have any stromal cells of the CC genotype. Therefore, the data obtained from subjects of CC genotype need to be taken with caution. The miRNA-520 levels shown in the Supplementary Figure were from a single patient of endometriosis. It is not clear whether all the endometriosis patients have such a high miR-520 expression level. Further studies to validate our finding are warranted.
In conclusion, SNP rs7201 at the 3′-UTR is at the miRNA-520g binding site. The risk C allele can reduce miRNA-520s silencing effect leading to increase MMP2 expression and endometriosis risk. From a microRNA pharmacogenomic point of view, a subject carrying the risk C allele may be more beneficial for the treatment by an MMP inhibitor.
References
Hadfield, R. M., Mardon, H. J., Barlow, D. H. & Kennedy, S. H. Endometriosis in monozygotic twins. Fertil. Steril. 68, 941–942 (1997).
Simpson, J. L. & Bischoff, F. Z. Heritability and molecular genetic studies of endometriosis. Ann. NY Acad. Sci. 955, 239–251 (2002).
Tempfer, C. B., Simoni, M., Destenaves, B. & Fauser, B. C. Functional genetic polymorphisms and female reproductive disorders: part II—endometriosis. Hum. Reprod. Update 15, 97–118 (2009).
Giudice, L. C. & Kao, L. C. Endometriosis. Lancet 364, 1789–1799 (2004).
Curry, T. E. & Osteen, K. G. The matrix metalloproteinase system: changes, regulation, and impact throughout the ovarian and uterine reproductive cycle. Endocr. Rev. 24, 428–465 (2003).
Chung, H. W., Lee, J. Y., Moon, H. S., Hur, S. E., Park, M. H., Wen, Y. et al. Matrix metalloproteinase-2, membranous type 1 matrix metalloproteinase, and tissue inhibitor of metalloproteinase-2 expression in ectopic and eutopic endometrium. Fertil. Steril. 78, 787–795 (2002).
Rawdanowicz, T. J., Hampton, A. L., Nagase, H., Woolley, D. E. & Salamonsen, L. A. Matrix metalloproteinase production by cultured human endometrial stromal cells: identification of interstitial collagenase, gelatinase-A, gelatinase-B, and stromelysin-1 and their differential regulation by interleukin-1 alpha and tumor necrosis factor-alpha. J. Clin. Endocrinol. Metab. 79, 530–536 (1994).
Huang, H. F., Hong, L. H., Tan, Y. & Sheng, J. Z. Matrix metalloproteinase 2 is associated with changes in steroid hormones in the sera and peritoneal fluid of patients with endometriosis. Fertil. Steril. 81, 1235–1239 (2004).
Inagaki, N., Ung, L., Otani, T., Wilkinson, D. & Lopata, A. Uterine cavity matrix metalloproteinases and cytokines in patients with leiomyoma, adenomyosis or endometrial polyp. Eur. J. Obstet. Gynecol. Reprod. Biol. 111, 197–203 (2003).
Uzan, C., Cortez, A., Dufournet, C., Fauvet, R., Siffroi, J. P. & Darai, E. Eutopic endometrium and peritoneal, ovarian and bowel endometriotic tissues express a different profile of matrix metalloproteinases-2, -3 and -11, and of tissue inhibitor metalloproteinases-1 and -2. Virchows Arch. 445, 603–609 (2004).
Juo, S. H., Wang, T. N., Lee, J. N., Wu, M. T., Long, C. Y. & Tsai, E. M. CYP17, CYP1A1 and COMT polymorphisms and the risk of adenomyosis and endometriosis in Taiwanese women. Hum. Reprod. 21, 1498–1502 (2006).
Tsai, H. T., Wu, C. H., Lai, H. L., Li, R. N., Tung, Y. C., Chuang, H. Y. et al. Association between quantitative high-risk human papillomavirus DNA load and cervical intraepithelial neoplasm risk. Cancer Epidemiol. Biomarkers Prev. 14, 1–6 (2005).
Wu, M. T., Lee, L. H., Ho, C. K., Liu, C. L., Wu, T. N., Wu, S. C. et al. Lifetime exposure to environmental tobacco smoke and cervical intraepithelial neoplasms among nonsmoking Taiwanese women. Arch. Environ. Health 58, 353–359 (2003).
Juo, S. H., Wu, R., Lin, C. S., Wu, M. T., Lee, J. N. & Tsai, E. M. A functional promoter polymorphism in interleukin-10 gene influences susceptibility to endometriosis. Fertil. Steril. 92, 1228–1233 (2009).
Price, S. J., Greaves, D. R. & Watkins, H. Identification of novel, functional genetic variants in the human matrix metalloproteinase-2 gene: role of Sp1 in allele-specific transcriptional regulation. J. Biol. Chem. 276, 7549–7558 (2001).
Wang, L. F., Chien, C. Y., Kuo, W. R., Tai, C. F. & Juo, S. H. Matrix metalloproteinase-2 gene polymorphisms in nasal polyps. Arch. Otolaryngol. Head Neck Surg. 134, 852–856 (2008).
Tzeng, J. Y., Wang, C. H., Kao, J. T. & Hsiao, C. K. Regression-based association analysis with clustered haplotypes through use of genotypes. Am. J. Hum. Genet. 78, 231–242 (2006).
Ueda, M., Yamashita, Y., Takehara, M., Terai, Y., Kumagai, K., Ueki, K. et al. Gene expression of adhesion molecules and matrix metalloproteinases in endometriosis. Gynecol. Endocrinol. 16, 391–402 (2002).
Lin, S. C., Lo, S. S., Liu, C. J., Chung, M. Y., Huang, J. W. & Chang, K. W. Functional genotype in matrix metalloproteinases-2 promoter is a risk factor for oral carcinogenesis. J. Oral Pathol. Med. 33, 405–409 (2004).
Ye, S. Influence of matrix metalloproteinase genotype on cardiovascular disease susceptibility and outcome. Cardiovasc. Res. 69, 636–645 (2006).
Saare, M., Lamp, M., Kaart, T., Karro, H., Kadastik, U., Metspalu, A. et al. Polymorphisms in MMP-2 and MMP-9 promoter regions are associated with endometriosis. Fertil. Steril. 94, 1560–1563 (2010).
Kang, S., Zhao, X. W., Wang, N., Chen, S. C., Zhou, R. M. & Li, Y. Association of polymorphisms of the MMP-2 and TIMP-2 genes with the risk of endometriosis in North Chinese women. Fertil. Steril. 90, 2023–2029 (2008).
Zhao, X. W., Li, Y., Wang, N., Zhao, J., Li, X. L., Liu, Q. et al. Study on the association of SNPs of MMP-2 and TIMP-2 genes with the risk of endometriosis and adenomyosis. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 25, 280–283 (2008).
Fatar, M., Stroick, M., Steffens, M., Senn, E., Reuter, B., Bukow, S. et al. Single-nucleotide polymorphisms of MMP-2 gene in stroke subtypes. Cerebrovasc. Dis. 26, 113–119 (2008).
Pan, Q., Luo, X., Toloubeydokhti, T. & Chegini, N. The expression profile of micro-RNA in endometrium and endometriosis and the influence of ovarian steroids on their expression. Mol. Hum. Reprod. 13, 797–806 (2007).
Ohlsson Teague, E. M., Van der Hoek, K. H., Van der Hoek, M. B., Perry, N., Wagaarachchi, P., Robertson, S. A. et al. MicroRNA-regulated pathways associated with endometriosis. Mol. Endocrinol. 23, 265–275 (2009).
Filigheddu, N., Gregnanin, I., Porporato, P. E., Surico, D., Perego, B., Galli, L. et al. Differential expression of microRNAs between eutopic and ectopic endometrium in ovarian endometriosis. J. Biomed. Biotechnol. 2010, 369549 (2010).
Hawkins, S. M., Creighton, C. J., Han, D. Y., Zariff, A., Anderson, M. L., Gunaratne, P. H. et al. Functional microRNA involved in endometriosis. Mol. Endocrinol. 25, 821–832 (2011).
Melichar, V. O., Behr-Roussel, D., Zabel, U., Uttenthal, L. O., Rodrigo, J., Rupin, A. et al. Reduced cGMP signaling associated with neointimal proliferation and vascular dysfunction in late-stage atherosclerosis. Proc. Natl Acad. Sci. USA 101, 16671–16676 (2004).
Mishra, P. J., Humeniuk, R., Longo-Sorbello, G. S., Banerjee, D. & Bertino, J. R. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc. Natl Acad. Sci. USA 104, 13513–13518 (2007).
Mishra, P. J., Banerjee, D. & Bertino, J. R. MiRSNPs or MiR-polymorphisms, new players in microRNA mediated regulation of the cell: introducing microRNA pharmacogenomics. Cell Cycle 7, 853–858 (2008).
Zhao, Z. Z., Croft, L., Nyholt, D. R., Chapman, B., Treloar, S. A., Hull, M. L. et al. Evaluation of polymorphisms in predicted target sites for micro RNAs differentially expressed in endometriosis. Mol. Hum. Reprod. 17, 92–103 (2011).
Grechukhina, O., Petracco, R., Popkhadze, S., Massasa, E., Paranjape, T., Chan, E. et al. A polymorphism in a let-7 microRNA binding site of KRAS in women with endometriosis. EMBO Mol. Med. 4, 206–217 (2012).
Yu, C., Pan, K., Xing, D., Liang, G., Tan, W., Zhang, L. et al. Correlation between a single nucleotide polymorphism in the matrix metalloproteinase-2 promoter and risk of lung cancer. Cancer Res. 62, 6430–6433 (2002).
Acknowledgements
This work was supported by the National Science Council, Taiwan (Grant numbers NSC 97-2314-B-037-010-MY3, 99-2628-B-037-009-MY3); Kaohsiung Medical University Research Foundation (KMUER-004) and the Kaohsiung Medical University Hospital Research Fund (Grant Numbers KMUH98-8I11, KMUH 99-9I04 and KMUH 99-9R30).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Tsai, EM., Wang, YS., Lin, CS. et al. A microRNA-520 mirSNP at the MMP2 gene influences susceptibility to endometriosis in Chinese women. J Hum Genet 58, 202–209 (2013). https://doi.org/10.1038/jhg.2013.1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2013.1
Keywords
This article is cited by
-
The effect of MicroRNAs variants on idiopathic recurrent pregnancy loss
Journal of Assisted Reproduction and Genetics (2023)
-
Circulating non-coding RNAs as non-invasive diagnostic markers of endometriosis: a comprehensive meta-analysis
Archives of Gynecology and Obstetrics (2019)