Abstract
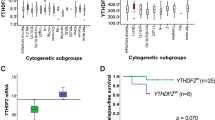
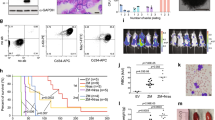
The mechanisms by which AML1/ETO (A/E) fusion protein induces leukemogenesis in acute myeloid leukemia (AML) without mutagenic events remain elusive. Here we show that interactions between A/E and hypoxia-inducible factor 1α (HIF1α) are sufficient to prime leukemia cells for subsequent aggressive growth. In agreement with this, HIF1α is highly expressed in A/E-positive AML patients and strongly predicts inferior outcomes, regardless of gene mutations. Co-expression of A/E and HIF1α in leukemia cells causes a higher cell proliferation rate in vitro and more serious leukemic status in mice. Mechanistically, A/E and HIF1α form a positive regulatory circuit and cooperate to transactivate DNMT3a gene leading to DNA hypermethylation. Pharmacological or genetic interventions in the A/E–HIF1α loop results in DNA hypomethylation, a re-expression of hypermethylated tumor-suppressor p15INK4b and the blockage of leukemia growth. Thus high HIF1α expression serves as a reliable marker, which identifies patients with a poor prognosis in an otherwise prognostically favorable AML group and represents an innovative therapeutic target in high-risk A/E-driven leukemia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Peterson LF, Zhang DE . The 8;21 translocation in leukemogenesis. Oncogene 2004; 23: 4255–4262.
Yuan Y, Zhou L, Miyamoto T, Iwasaki H, Harakawa N, Hetherington CJ et al. AML1-ETO expression is directly involved in the development of acute myeloid leukemia in the presence of additional mutations. Proc Natl Acad Sci USA 2001; 98: 10398–10403.
Wang YY, Zhou GB, Yin T, Chen B, Shi JY, Liang WX et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: implication in stepwise leukemogenesis and response to Gleevec. Proc Natl Acad Sci USA 2005; 102: 1104–1109.
Schessl C, Rawat VP, Cusan M, Deshpande A, Kohl TM, Rosten PM et al. The AML1-ETO fusion gene and the FLT3 length mutation collaborate in inducing acute leukemia in mice. J Clin Invest 2005; 115: 2159–2168.
Schneider F, Bohlander SK, Schneider S, Papadaki C, Kakadyia P, Dufour A et al. AML1-ETO meets JAK2: clinical evidence for the two hit model of leukemogenesis from a myeloproliferative syndrome progressing to acute myeloid leukemia. Leukemia 2007; 21: 2199–2201.
Peterson LF, Boyapati A, Ahn EY, Biggs JR, Okumura AJ, Lo MC et al. Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts. Blood 2007; 110: 799–805.
Kuchenbauer F, Schnittger S, Look T, Gilliland G, Tenen D, Haferlach T et al. Identification of additional cytogenetic and molecular genetic abnormalities in acute myeloid leukaemia with t(8;21)/AML1-ETO. Br J Haematol 2006; 134: 616–619.
Dohner K, Du J, Corbacioglu A, Scholl C, Schlenk RF, Dohner H . JAK2V617F mutations as cooperative genetic lesions in t(8;21)-positive acute myeloid leukemia. Haematologica 2006; 91: 1569–1570.
Paschka P, Marcucci G, Ruppert AS, Mrozek K, Chen H, Kittles RA et al. Adverse prognostic significance of KIT mutations in adult acute myeloid leukemia with inv(16) and t(8;21): a Cancer and Leukemia Group B Study. J Clin Oncol 2006; 24: 3904–3911.
Keith B, Simon MC . Hypoxia-inducible factors, stem cells, and cancer. Cell 2007; 129: 465–472.
Stroka DM, Burkhardt T, Desbaillets I, Wenger RH, Neil DA, Bauer C et al. HIF-1 is expressed in normoxic tissue and displays an organ-specific regulation under systemic hypoxia. FASEB J 2001; 15: 2445–2453.
Kuschel A, Simon P, Tug S . Functional regulation of HIF-1alpha under normoxia—is there more than post-translational regulation? J Cell Physiol 2012; 227: 514–524.
Gerber SA, Pober JS . IFN-alpha induces transcription of hypoxia-inducible factor-1alpha to inhibit proliferation of human endothelial cells. J Immunol 2008; 181: 1052–1062.
Wang Y, Liu Y, Malek SN, Zheng P . Targeting HIF1alpha eliminates cancer stem cells in hematological malignancies. Cell Stem Cell 2011; 8: 399–411.
Matsunaga T, Imataki O, Torii E, Kameda T, Shide K, Shimoda H et al. Elevated HIF-1alpha expression of acute myelogenous leukemia stem cells in the endosteal hypoxic zone may be a cause of minimal residual disease in bone marrow after chemotherapy. Leuk Res 2012; 36: e122–e124.
Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer cell 2010; 17: 13–27.
Linggi B, Muller-Tidow C, van de Locht L, Hu M, Nip J, Serve H et al. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med 2002; 8: 743–750.
Kitabayashi I, Ida K, Morohoshi F, Yokoyama A, Mitsuhashi N, Shimizu K et al. The AML1-MTG8 leukemic fusion protein forms a complex with a novel member of the MTG8(ETO/CDR) family, MTGR1. Mol Cell Biol 1998; 18: 846–858.
Wang J, Hoshino T, Redner RL, Kajigaya S, Liu JM . ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA 1998; 95: 10860–10865.
Liu S, Shen T, Huynh L, Klisovic MI, Rush LJ, Ford JL et al. Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia. Cancer Res 2005; 65: 1277–1284.
Liu Y, Cheney MD, Gaudet JJ, Chruszcz M, Lukasik SM, Sugiyama D et al. The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO's activity. Cancer cell 2006; 9: 249–260.
Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer cell 2007; 12: 457–466.
Liu S, Wu LC, Pang J, Santhanam R, Schwind S, Wu YZ et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer cell 2010; 17: 333–347.
Shen N, Yan F, Pang J, Wu LC, Al-Kali A, Litzow MR et al. A nucleolin-DNMT1 regulatory axis in acute myeloid leukemogenesis. Oncotarget 2014; 5: 5494–5509.
de Jonge HJ, Valk PJ, Veeger NJ, ter Elst A, den Boer M L, Cloos J et al. High VEGFC expression is associated with unique gene expression profiles and predicts adverse prognosis in pediatric and adult acute myeloid leukemia. Blood 2010; 116: 1747–1754.
Litz J, Krystal GW . Imatinib inhibits c-Kit-induced hypoxia-inducible factor-1alpha activity and vascular endothelial growth factor expression in small cell lung cancer cells. Mol Cancer Ther 2006; 5: 1415–1422.
Gardini A, Cesaroni M, Luzi L, Okumura AJ, Biggs JR, Minardi SP et al. AML1/ETO oncoprotein is directed to AML1 binding regions and co-localizes with AML1 and HEB on its targets. PLoS Genet 2008; 4: e1000275.
Ptasinska A, Assi SA, Mannari D, James SR, Williamson D, Dunne J et al. Depletion of RUNX1/ETO in t(8;21) AML cells leads to genome-wide changes in chromatin structure and transcription factor binding. Leukemia 2012; 26: 1829–1841.
Ortiz-Barahona A, Villar D, Pescador N, Amigo J, del Peso L . Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res 2010; 38: 2332–2345.
Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci USA 2009; 106: 4260–4265.
Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J et al. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem 2009; 284: 16767–16775.
Yan M, Kanbe E, Peterson LF, Boyapati A, Miao Y, Wang Y et al. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat Med 2006; 12: 945–949.
Kong D, Park EJ, Stephen AG, Calvani M, Cardellina JH, Monks A et al. Echinomycin, a small-molecule inhibitor of hypoxia-inducible factor-1 DNA-binding activity. Cancer Res 2005; 65: 9047–9055.
Herman JG, Jen J, Merlo A, Baylin SB . Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res 1996; 56: 722–727.
Higuchi M, O'Brien D, Kumaravelu P, Lenny N, Yeoh EJ, Downing JR . Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8; 21) acute myeloid leukemia. Cancer cell 2002; 1: 63–74.
Vaupel P . The role of hypoxia-induced factors in tumor progression. Oncologist 2004; 9 (Suppl 5): 10–17.
Unruh A, Ressel A, Mohamed HG, Johnson RS, Nadrowitz R, Richter E et al. The hypoxia-inducible factor-1 alpha is a negative factor for tumor therapy. Oncogene 2003; 22: 3213–3220.
Baba Y, Nosho K, Shima K, Irahara N, Chan AT, Meyerhardt JA et al. HIF1A overexpression is associated with poor prognosis in a cohort of 731 colorectal cancers. Am J Pathol 2010; 176: 2292–2301.
Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood 1998; 92: 2322–2333.
Cairoli R, Beghini A, Grillo G, Nadali G, Elice F, Ripamonti CB et al. Prognostic impact of c-KIT mutations in core binding factor leukemias: an Italian retrospective study. Blood 2006; 107: 3463–3468.
Yoo SJ, Chi HS, Jang S, Seo EJ, Seo JJ, Lee JH et al. Quantification of AML1-ETO fusion transcript as a prognostic indicator in acute myeloid leukemia. Haematologica 2005; 90: 1493–1501.
Li Y, Gao L, Luo X, Wang L, Gao X, Wang W et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8; 21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood 2013; 121: 499–509.
Maiques-Diaz A, Chou FS, Wunderlich M, Gomez-Lopez G, Jacinto FV, Rodriguez-Perales S et al. Chromatin modifications induced by the AML1-ETO fusion protein reversibly silence its genomic targets through AML1 and Sp1 binding motifs. Leukemia 2012; 26: 1329–1337.
Wang L, Gural A, Sun XJ, Zhao X, Perna F, Huang G et al. The leukemogenicity of AML1-ETO is dependent on site-specific lysine acetylation. Science 2011; 333: 765–769.
Shia WJ, Okumura AJ, Yan M, Sarkeshik A, Lo MC, Matsuura S et al. PRMT1 interacts with AML1-ETO to promote its transcriptional activation and progenitor cell proliferative potential. Blood 2012; 119: 4953–4962.
Peng ZG, Zhou MY, Huang Y, Qiu JH, Wang LS, Liao SH et al. Physical and functional interaction of Runt-related protein 1 with hypoxia-inducible factor-1alpha. Oncogene 2008; 27: 839–847.
Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G et al. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci USA 2002; 99: 5367–5372.
Fahling M, Persson AB, Klinger B, Benko E, Steege A, Kasim M et al. Multilevel regulation of HIF-1 signaling by TTP. Mol Biol Cell 2012; 23: 4129–4141.
Lee JS, Kim Y, Bhin J, Shin HJ, Nam HJ, Lee SH et al. Hypoxia-induced methylation of a pontin chromatin remodeling factor. Proc Natl Acad Sci USA 2011; 108: 13510–13515.
Robinson CM, Neary R, Levendale A, Watson CJ, Baugh JA . Hypoxia-induced DNA hypermethylation in human pulmonary fibroblasts is associated with Thy-1 promoter methylation and the development of a pro-fibrotic phenotype. Respir Res 2012; 13: 74.
Shahrzad S, Bertrand K, Minhas K, Coomber BL . Induction of DNA hypomethylation by tumor hypoxia. Epigenetics 2007; 2: 119–125.
Liu Q, Liu L, Zhao Y, Zhang J, Wang D, Chen J et al. Hypoxia induces genomic DNA demethylation through the activation of HIF-1alpha and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther 2011; 10: 1113–1123.
Acknowledgements
This work was partially supported by grants from the National Institutes of Health/National Cancer Institute (R01CA149623, R01CA104509 and R21CA155915), the Hormel Foundation, the National Natural Science Foundation of China (90919044, 30971297, 81170518, 81000221, 81370010, 81171820 and 81370635), the National Public Health Grand Research Foundation (201202017), the Capital Public Health Project (Z111107067311070), the Capital Medical Development Scientific Research Fund (2007–2040), the Beijing Natural Science Foundation (7122169 and 7112126), the Beijing New Stars Program of Science and Technology (2010B075) and the Italian Association for Cancer Research (IG-11949).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Gao, X., Yan, F., Lin, J. et al. AML1/ETO cooperates with HIF1α to promote leukemogenesis through DNMT3a transactivation. Leukemia 29, 1730–1740 (2015). https://doi.org/10.1038/leu.2015.56
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2015.56
This article is cited by
-
The chromatin architectural regulator SND1 mediates metastasis in triple-negative breast cancer by promoting CDH1 gene methylation
Breast Cancer Research (2023)
-
Crosstalk between DNA methylation and hypoxia in acute myeloid leukaemia
Clinical Epigenetics (2023)
-
HIF1α-mediated transactivation of WTAP promotes AML cell proliferation via m6A-dependent stabilization of KDM4B mRNA
Leukemia (2023)
-
Aberrant DNA methylation in t(8;21) acute myeloid leukemia
Genome Instability & Disease (2022)
-
Novel insights into roles of N6-methyladenosine reader YTHDF2 in cancer progression
Journal of Cancer Research and Clinical Oncology (2022)