Abstract
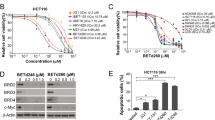
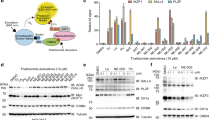
Bromodomain extraterminal protein (BETP) inhibitors transcriptionally repress oncoproteins and nuclear factor-κB (NF-κB) target genes that undermines the growth and survival of mantle cell lymphoma (MCL) cells. However, BET bromodomain inhibitor (BETi) treatment causes accumulation of BETPs, associated with reversible binding and incomplete inhibition of BRD4 that potentially compromises the activity of BETi in MCL cells. Unlike BETi, BET-PROTACs (proteolysis-targeting chimera) ARV-825 and ARV-771 (Arvinas, Inc.) recruit and utilize an E3-ubiquitin ligase to effectively degrade BETPs in MCL cells. BET-PROTACs induce more apoptosis than BETi of MCL cells, including those resistant to ibrutinib. BET-PROTAC treatment induced more perturbations in the mRNA and protein expressions than BETi, with depletion of c-Myc, CDK4, cyclin D1 and the NF-κB transcriptional targets Bcl-xL, XIAP and BTK, while inducing the levels of HEXIM1, NOXA and CDKN1A/p21. Treatment with ARV-771, which possesses superior pharmacological properties compared with ARV-825, inhibited the in vivo growth and induced greater survival improvement than the BETi OTX015 of immune-depleted mice engrafted with MCL cells. Cotreatment of ARV-771 with ibrutinib or the BCL2 antagonist venetoclax or CDK4/6 inhibitor palbociclib synergistically induced apoptosis of MCL cells. These studies highlight promising and superior preclinical activity of BET-PROTAC than BETi, requiring further in vivo evaluation of BET-PROTAC as a therapy for ibrutinib-sensitive or -resistant MCL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Zhang J, Jima D, Moffitt AB, Liu Q, Czader M, Hsi ED et al. The genomic landscape of mantle cell lymphoma is related to the epigenetically determined chromatin state of normal B cells. Blood 2014; 123: 2988–2996.
Jares P, Colomer D, Campo E . Molecular pathogenesis of mantle cell lymphoma. J Clin Invest 2012; 122: 3416–3423.
Beà S, Valdés-Mas R, Navarro A, Salaverria I, Martín-Garcia D, Jares P et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci USA 2013; 110: 18250–18255.
Parekh S, Weniger MA, Wiestner A . New molecular targets in mantle cell lymphoma. Semin Cancer Biol 2011; 21: 335–346.
Young RM, Staudt LM . Targeting pathological B cell receptor signaling in lymphoid malignancies. Nat Rev Drug Discov 2013; 12: 229–243.
Perez-Galan P, Dreyling M, Wiestner A . Mantle cell lymphoma: biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood 2011; 117: 26–38.
Campo E, Rule S . Mantle cell lymphoma: evolving management strategies. Blood 2015; 125: 48–55.
Wang ML, Rule S, Martin P, Goy A, Auer R, Kahl BS et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med 2013; 369: 507–516.
Herrera A, Jacobsen ED . Ibrutinib for the treatment of mantle cell lymphoma. Clin Cancer Res 2014; 20: 5365–5371.
Wang ML, Blum KA, Martin P, Goy A, Auer R, Kahl BS et al. Long-term follow-up of MCL patients treated with single-agent ibrutinib: updated safety and efficacy results. Blood 2015; 126: 739–745.
Rahal R, Frick M, Romero R, Korn JM, Kridel R, Chan FC et al. Pharmacological and genomic profiling identifies NF-κB-targeted treatment strategies for mantle cell lymphoma. Nat Med 2014; 20: 87–92.
Colomer D, Campo E . Unlocking new therapeutic targets and resistance mechanisms in mantle cell lymphoma. Cancer Cell 2014; 25: 7–9.
Jacobson C, Kopp N, Layer JV . HSP90 inhibition overcomes ibrutinib resistance in mantle cell lymphoma. Blood 2016; 128: 2517–2526.
Sun B, Shah B, Fiskus W, Qi J, Rajapakshe K, Coarfa C et al. Synergistic activity of BET protein antagonist-based combinations in mantle cell lymphoma cells sensitive or resistant to ibrutinib. Blood 2015; 126: 1565–1574.
Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S et al. DRUG DEVELOPMENT. Phthalidimide conjugation as a strategy for in vivo target protein degradation. Science 2015; 348: 1376–1381.
Vallabhapurapu S, Karin M . Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 2009; 27: 693–733.
Hayden MS, Ghosh S . Shared principles in NF-kappaB signaling. Cell 2008; 132: 344–362.
Dang CV . MYC on the path to cancer. Cell 2012; 149: 22–35.
Lu J, Qian Y, Altieri M, Dong H, Wang J, Raina K et al. Hijacking the E3 ubiquitin ligase cereblon to efficiently target BRD4. Chem Biol 2015; 22: 755–763.
Toure M, Crews CM . Small-molecule PROTACS: new approaches to protein degradation. Agnew Chem Int Ed Engl 2016; 55: 1966–1973.
Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci USA 2016; 113: 7124–7129.
Boi M, Gaudio E, Bonetti P, Kwee I, Bernasconi E, Tarantelli C et al. The inhibitor OTX015 affects pathogenetic pathways in preclinical B-cell tumor models and synergizes with targeted drugs. Clin Cancer Res 2015; 21: 1628–1638.
Eichner R, Heider M, Fernández-Sáiz V, van Bebber F, Garz AK, Lemeer S et al. Immunomodulatory drugs disrupt the cereblon-CD147-MCT1 axis to exert antitumor activity and teratogenicity. Nat Med 2016; 22: 735–743.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B . Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008; 5: 621–628.
Chou TC, Talalay P . Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 1984; 22: 27–55.
Saenz DT, Fiskus W, Qian Y, Manshouri T, Rajapakshe K, Raina K et al. Novel BET protein proteolysis targeting chimera (BET-PROTAC) exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm (MPN) secondary (s) AML cells. Leukemia 2017. e-pub ahead of print 31 January 2017, doi:10.1038/leu.2016.393.
Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010; 141: 69–80.
Devaraj SG, Fiskus W, Shah B, Qi J, Sun B, Iyer SP et al. HEXIM1 induction is mechanistically involved in mediating anti-AML activity of BET protein bromodomain antagonist. Leukemia 2016; 30: 504–508.
Saenz DT, Fiskus W, Manshouri T, Rajapakshe K, Krieger S, Sun B et al. BET protein bromodomain inhibitor-based combinations are highly active against post-myeloproliferative neoplasm secondary AML cells. Leukemia 2017; 31: 678–687.
Kornblau SM, Tibes R, Qiu YH, Chen W, Kantarjian HM, Andreeff M et al. Functional proteomic profiling of AML predicts response and survival. Blood 2009; 113: 154–164.
Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J et al. Brd4 maintains constitutively active NF-kappaB in cancer cells by binding to acetylated RelA. Oncogene 2014; 33: 2395–2404.
Huang B, Yang XD, Zhou MM, Ozato K, Chen LF . Brd4 coactivates transcriptional activation of NF-kappaB via specific binding to acetylated RelA. Mol Cell Biol 2009; 29: 1375–1387.
Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR . BET Bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol Cell 2015; 58: 1028–1039.
Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol 2014; 21: 1047–1057.
Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA et al. Super enhancers in the control of cell identity and disease. Cell 2013; 155: 934–947.
Shi J, Vakoc CR . The mechanism behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 2014; 54: 728–736.
Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153: 320–334.
Shen C, Ipsaro JJ, Shi J, Milazzo JP, Wang E, Roe JS et al. NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell 2015; 60: 847–859.
Bhagwat AS, Roe JS, Mok BY, Hohmann AF, Shi J, Vakoc CR . BET bromodomain inhibition releases the mediator complex from select cis-regulatory elements. Cell Rep 2016; 15: 519–530.
Itzen F, Greifenberg AK, Bosken CA, Geyer M . Brd4 activates P-TEFB for RNA polymerase II CTD phosphorylation. Nucleic Acids Res 2014; 42: 7577–7590.
Nechaev S, Adelman K . Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta 2011; 1809: 34–45.
Levine M, Cattoglio C, Tjian R . Looping back to leap forward: transcription enters a new era. Cell 2014; 157: 13–25.
Lee TI, Young RA . Transcriptional regulation and its misregulation in disease. Cell 2013; 152: 1237–1251.
Ma J, Lu P, Guo A, Cheng S, Zong H, Martin P et al. Characterization of ibrutinib-sensitive and -resistant mantle lymphoma cells. Br J Haematol 2014; 166: 849–861.
Chiron D, Di Liberto M, Martin P, Huang X, Sharman J, Blecua P et al. Cell-cycle reprogramming for PI3K inhibition overrides a relapse-specific C481S BTK mutation revealed by longitudinal functional genomics in mantle cell lymphoma. Cancer Discov 2014; 4: 1022–1035.
Li Y, Bouchlaka MN, Wolff J, Grindle KM, Lu L, Qian S et al. FBXO10 deficiency and BTK activation upregulate BCL2 expression in mantle cell lymphoma. Oncogene 2016; 35: 6223–6234.
Chiron D, Bellanger C, Papin A, Tessoulin B, Dousset C, Maiga S et al. Rational targeted therapies to overcome microenvironment-dependent expansion of mantle cell lymphoma. Blood 2016; 128: 2808–2818.
Saba NS, Liu D, Herman SE, Underbayev C, Tian X, Behrend D et al. Pathogenic role of B-cell receptor signaling and canonical NF-κB activation in mantle cell lymphoma. Blood 2016; 128: 82–92.
Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 2013; 19: 202–208.
Leonard JP, LaCasce AS, Smith MR, Noy A, Chirieac LR, Rodig SJ et al. Selective CDK4/6 inhibition with tumor responses by PD0332991 in patients with mantle cell lymphoma. Blood 2012; 119: 4597–4607.
Rathert P, Roth M, Neumann T, Muerdter F, Roe JS, Muhar M et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature 2015; 525: 543–547.
Fong CY, Gilan O, Lam EY, Rubin AF, Ftouni S, Tyler D et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature 2015; 525: 538–542.
Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 2016; 529: 413–417.
Acknowledgements
We thank the Flow Cytometry and Cellular Imaging (FCCI) Core Facility and the Functional Proteomics RPPA Core facility that are supported by MD Anderson Cancer Center Support Grant 5P30 CA016672-40. The RPPA heatmaps were developed by the MD Anderson Cancer Center Department of Bioinformatics and Computational Biology, In Silico Solutions, Santeon and SRA International. This work was supported in part by US National Cancer Institute (NCI; MD Anderson TCGA Genome Data Analysis Center) Grant Numbers CA143883 and CA083639, the Mary K Chapman Foundation, the Michael & Susan Dell Foundation (honoring Lorraine Dell) and MD Anderson Cancer Center Support Grant P30 CA016672 (the Bioinformatics Shared Resource). This project was partially supported by CPRIT RP170295 (to CC), the shared Proteomics and Metabolomics core at Baylor College of Medicine with funding from the NIH (P30 CA125123), CPRIT Proteomics and Metabolomics Core Facility RP120092 (to KR and CC) and the NCI-recognized Dan L Duncan Cancer Center. CMC acknowledges support from the National Institutes of Health (Grant Number R35 CA197589).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
CMC is the founder and Chief Scientific Advisor of, and possesses an equity ownership stake in, Arvinas, Inc. YQ, KR, KGC, APC and AS are Arvinas employees and possess an equity ownership stake in Arvinas. The other authors state declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Supplementary information
Rights and permissions
About this article
Cite this article
Sun, B., Fiskus, W., Qian, Y. et al. BET protein proteolysis targeting chimera (PROTAC) exerts potent lethal activity against mantle cell lymphoma cells. Leukemia 32, 343–352 (2018). https://doi.org/10.1038/leu.2017.207
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2017.207
This article is cited by
-
Beyond canonical PROTAC: biological targeted protein degradation (bioTPD)
Biomaterials Research (2023)
-
Epigenetic regulation in hematopoiesis and its implications in the targeted therapy of hematologic malignancies
Signal Transduction and Targeted Therapy (2023)
-
Bromodomain and extraterminal (BET) proteins: biological functions, diseases, and targeted therapy
Signal Transduction and Targeted Therapy (2023)
-
The crosstalk between ubiquitination and endocrine therapy
Journal of Molecular Medicine (2023)
-
Tipping the balance: toward rational combination therapies to overcome venetoclax resistance in mantle cell lymphoma
Leukemia (2022)