Abstract
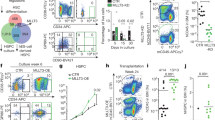
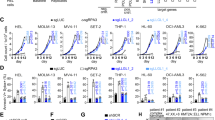
The balance between self-renewal and differentiation of adult stem cells is essential for tissue homeostasis. Here we show that in the haematopoietic system this process is governed by polycomb chromobox (Cbx) proteins. Cbx7 is specifically expressed in haematopoietic stem cells (HSCs), and its overexpression enhances self-renewal and induces leukaemia. This effect is dependent on integration into polycomb repressive complex-1 (PRC1) and requires H3K27me3 binding. In contrast, overexpression of Cbx2, Cbx4 or Cbx8 results in differentiation and exhaustion of HSCs. ChIP-sequencing analysis shows that Cbx7 and Cbx8 share most of their targets; we identified approximately 200 differential targets. Whereas genes targeted by Cbx8 are highly expressed in HSCs and become repressed in progenitors, Cbx7 targets show the opposite expression pattern. Thus, Cbx7 preserves HSC self-renewal by repressing progenitor-specific genes. Taken together, the presence of distinct Cbx proteins confers target selectivity to PRC1 and provides a molecular balance between self-renewal and differentiation of HSCs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Lewis, E. B. A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 (1978).
Simon, J. A. & Kingston, R. E. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 (2009).
Morey, L. & Helin, K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 35, 323–332 (2010).
Lessard, J., Baban, S. & Sauvageau, G. Stage-specific expression of polycomb group genes in human BM cells. Blood 91, 1216–1224 (1998).
Gunster, M. J. et al. Differential expression of human Polycomb groupproteins in various tissues and cell types. J. Cell. Biochem. Suppl. 36, 129–143 (2001).
Iwama, A. et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity 21, 843–851 (2004).
Kato, Y., Koseki, H., Vidal, M., Nakauchi, H. & Iwama, A. Unique composition of polycomb repressive complex 1 in hematopoietic stem cells. Int. J. Hematol. 85, 179–181 (2007).
Morey, L. et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 10, 47–62 (2012).
Tavares, L. et al. RYBP-PRC1 complexes mediate H2A ubiquitylation atpolycomb target sites independently of PRC2 and H3K27me3. Cell 148, 664–678 (2012).
O’Loghlen, A. et al. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell 10, 33–46 (2012).
Gao, Z. et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol. Cell 45, 344–356 (2012).
Maertens, G. N. et al. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS One 4, e6380 (2009).
Vandamme, J., Volkel, P., Rosnoblet, C., Le Faou, P. & Angrand, P. O. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol. Cell Proteomics 10, M110.002642 (2011).
Min, J., Zhang, Y. & Xu, R. M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17, 1823–1828 (2003).
Fischle, W. et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17, 1870–1881 (2003).
Bernstein, E. et al. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell Biol. 26, 2560–2569 (2006).
Kaustov, L. et al. Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 286, 521–529 (2011).
Van Lohuizen, M. et al. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65, 737–752 (1991).
Kamminga, L. M. et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood 107, 2170–2179 (2006).
Lessard, J. et al. Functional antagonism of the Polycomb-Group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev. 13, 2691–2703 (1999).
Park, I. K. et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302–305 (2003).
Rizo, A., Dontje, B., Vellenga, E., de Haan, G. & Schuringa, J. J. Long-term maintenance of human hematopoietic stem/progenitor cells by expression of BMI1. Blood 111, 2621–2630 (2008).
Dephoure, N. et al. A quantitative atlas of mitotic phosphorylation. Proc. Natl Acad. Sci. USA 105, 10762–10767 (2008).
Scott, C. L. et al. Role of the chromobox protein CBX7 in lymphomagenesis. Proc. Natl Acad. Sci. USA 104, 5389–5394 (2007).
Ploemacher, R. E., van der Sluijs, J. P., van Beurden, C. A., Baert, M. R. & Chan, P. L. Use of limiting-dilution type long-term marrow cultures in frequency analysis of marrow-repopulating and spleen colony-forming hematopoietic stem cells in the mouse. Blood 78, 2527–2533 (1991).
De Haan, G., Nijhof, W. & Van Zant, G. Mouse strain-dependent changes in frequency and proliferation of hematopoietic stem cells during aging: correlation between lifespan and cycling activity. Blood 89, 1543–1550 (1997).
Gil, J., Bernard, D., Martinez, D. & Beach, D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat. Cell Biol. 6, 67–72 (2004).
Jacobs, J. J., Kieboom, K., Marino, S., DePinho, R. A. & van Lohuizen, M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397, 164–168 (1999).
Oguro, H. et al. Differential impact of Ink4a and Arf on hematopoietic stem cells and their BM microenvironment in Bmi1-deficient mice. J. Exp. Med. 203, 2247–2253 (2006).
Voy, B. H. et al. Extracting gene networks for low-dose radiation using graph theoretical algorithms. PLoS Comput. Biol. 2, e89 (2006).
Walasek, M. A. et al. The combination of valproic acid and lithiumdelays hematopoietic stem/progenitor cell differentiation. Blood 119, 3050–3059 (2012).
Colombo, E., Alcalay, M. & Pelicci, P. G. Nucleophosmin and its complex network: a possible therapeutic target in hematological diseases. Oncogene 30, 2595–2609 (2011).
Falini, B. et al. Acute myeloid leukemia with mutated nucleophosmin (NPM1): any hope for a targeted therapy? Blood Rev. 25, 247–254 (2011).
Yin, B. et al. A retroviral mutagenesis screen reveals strong cooperation between Bcl11a overexpression and loss of the Nf1 tumor suppressor gene. Blood 113, 1075–1085 (2009).
Deambrogi, C. et al. Analysis of the REL, BCL11A, and MYCN proto-oncogenes belonging to the 2p amplicon in chronic lymphocytic leukemia. Am. J. Hematol. 85, 541–544 (2010).
Osawa, M. et al. Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood 100, 2769–2777 (2002).
Laurent, B. et al. High-mobility group protein HMGB2 regulates human erythroid differentiation through trans-activation of GFI1B transcription. Blood 115, 687–695 (2010).
Van der Meer, L. T., Jansen, J. H. & van der Reijden, B. A. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia 24, 1834–1843 (2010).
Luis, N. M. et al. Regulation of human epidermal stem cell proliferation and senescence requires polycomb- dependent and -independent functions of Cbx4. Cell Stem Cell 9, 233–246 (2011).
Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. Int. AAAI Conf. on Weblogs and Social Media (2009).
Dykstra, B., Olthof, S., Schreuder, J., Ritsema, M. & de Haan, G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 208, 2691–2703 (2011).
Challen, G. A., Boles, N., Lin, K. K. & Goodell, M. A. Mouse hematopoietic stem cell identification and analysis. Cytometry A 75, 14–24 (2009).
Dietrich, N. et al. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J. 26, 1637–1648 (2007).
Gerrits, A. et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood 115, 2610–2618 (2010).
Moffat, J. et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 (2006).
Sarbassov, D. D., Guertin, D. A., Ali, S. M. & Sabatini, D. M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005).
Van Os, R. P., Dethmers-Ausema, B. & de Haan, G. In vitro assays forcobblestone area-forming cells, LTC-IC, and CFU-C. Methods Mol. Biol. 430, 143–157 (2008).
Frank, S. R., Schroeder, M., Fernandez, P., Taubert, S. & Amati, B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15, 2069–2082 (2001).
Bracken, A. P., Dietrich, N., Pasini, D., Hansen, K. H. & Helin, K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136 (2006).
Dietrich, N. et al. REST-mediated recruitment of polycomb repressor complexes in mammalian cells. PLoS Genet. 8, e1002494 (2012).
Bracken, A. P. et al. The Polycomb group proteins bind throughout the INK4a-ARF locus and are disassociated in senescent cells. Genes Dev. 21, 525–530 (2007).
Giardine, B. et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455 (2005).
Blankenberg, D. et al. Manipulation of FASTQ data with Galaxy. Bioinformatics 26, 1783–1785 (2010).
Goecks, J., Nekrutenko, A. & Taylor, J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86 (2010).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Feng, J., Liu, T. & Zhang, Y. Using MACS to identify peaks from ChIP-Seq data. Curr. Protoc. Bioinform. (2011) Chapter 2, Unit 2 14.
Zhu, L. J. et al. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinform. 11, 237 (2010).
Durinck, S., Spellman, P. T., Birney, E. & Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184–1191 (2009).
de St Groth, Fazekas The evaluation of limiting dilution assays. J. Immunol. Methods 49, R11–R23 (1982).
Acknowledgements
We thank H. Moes, G. Mesander, H. de Bruin and R. J. van der Lei for expert flow cytometry assistance, the entire staff of the Central Animal Facility at the UMCG, K. Magnussen from the National High-throughput Sequencing Centre of the University of Copenhagen, B. Dethmers-Ausema, R. Bron, F. Feringa, K. van der Laan, V. Stojanovska and K. Wakker for laboratory assistance, J. Engelbert for bioinformatical assistance, and B. Dykstra, M. Niemantsverdriet, R. van Os and H. Schepers for valuable scientific discussions. We also acknowledge financial support from the Netherlands Organization for Scientific Research (VICI 918-76-601 and ALW to G.d.H), the Netherlands Institute for Regenerative Medicine, the Dutch Cancer Society (grant 2007-3729, and UMCG-2011-5277 to S.B.) and the European Community (EuroSystem, 200720). The work in the Helin laboratory was supported by the Danish National Research Foundation, the Novo Nordisk Foundation and the Danish Cancer Society.
Author information
Authors and Affiliations
Contributions
K.K., L.B., K.H. and G.d.H. initiated research and developed the concept of the paper. K.K., L.B., and G.d.H. designed research; K.K. and V.R. performed experiments with contributions from M.B., E.W., S.O., M.R., S.B. and X.W; E.Z. performed bioinformatics analyses with contributions from L.B. and K.K.; K.K. and L.B. analysed and interpreted data; and K.K. wrote the manuscript with contributions from L.B., K.H. and G.d.H.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1136 kb)
Supplementary Table 1
Supplementary Information (XLS 55 kb)
Supplementary Table 2
Supplementary Information (XLS 41 kb)
Supplementary Table 3
Supplementary Information (XLS 32 kb)
Supplementary Table 4
Supplementary Information (XLS 149 kb)
Supplementary Table 5
Supplementary Information (XLSX 109 kb)
Supplementary Table 6
Supplementary Information (XLSX 20 kb)
Supplementary Table 7
Supplementary Information (XLSX 37 kb)
Supplementary Table 8
Supplementary Information (XLS 3193 kb)
Rights and permissions
About this article
Cite this article
Klauke, K., Radulović, V., Broekhuis, M. et al. Polycomb Cbx family members mediate the balance between haematopoietic stem cell self-renewal and differentiation. Nat Cell Biol 15, 353–362 (2013). https://doi.org/10.1038/ncb2701
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2701
This article is cited by
-
E3 ubiquitin ligase on the biological properties of hematopoietic stem cell
Journal of Molecular Medicine (2023)
-
St18 specifies globus pallidus projection neuron identity in MGE lineage
Nature Communications (2022)
-
Context-specific Polycomb mechanisms in development
Nature Reviews Genetics (2022)
-
Liver-specific deletion of miR-181ab1 reduces liver tumour progression via upregulation of CBX7
Cellular and Molecular Life Sciences (2022)
-
AKT-mediated regulation of chromatin ubiquitylation and tumorigenesis through Mel18 phosphorylation
Oncogene (2021)