Abstract
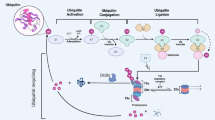
The tumour suppressor PTEN is frequently lost in human cancers. In addition to gene mutations and deletions, recent studies have revealed the importance of post-translational modifications, such as ubiquitylation, in the regulation of PTEN stability, activity and localization. However, the deubiquitylase that regulates PTEN polyubiquitylation and protein stability remains unknown. Here we screened a total of 30 deubiquitylating enzymes (DUBs) and identified five DUBs that physically associate with PTEN. One of them, USP13, stabilizes the PTEN protein through direct binding and deubiquitylation of PTEN. Loss of USP13 in breast cancer cells promotes AKT phosphorylation, cell proliferation, anchorage-independent growth, glycolysis and tumour growth through downregulation of PTEN. Conversely, overexpression of USP13 suppresses tumorigenesis and glycolysis in PTEN-positive but not PTEN-null breast cancer cells. Importantly, USP13 protein is downregulated in human breast tumours and correlates with PTEN protein levels. These findings identify USP13 as a tumour-suppressing protein that functions through deubiquitylation and stabilization of PTEN.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Leevers, S. J., Vanhaesebroeck, B. & Waterfield, M. D. Signalling through phosphoinositide 3-kinases: the lipids take centre stage. Curr. Opin. Cell Biol. 11, 219–225 (1999).
Maehama, T. & Dixon, J. E. The tumour suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 273, 13375–13378 (1998).
Di Cristofano, A., Pesce, B., Cordon-Cardo, C. & Pandolfi, P. P. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19, 348–355 (1998).
Garcia-Cao, I. et al. Systemic elevation of PTEN induces a tumour-suppressive metabolic state. Cell 149, 49–62 (2012).
Song, M. S., Salmena, L. & Pandolfi, P. P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 13, 283–296 (2012).
Liaw, D. et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat. Genet. 16, 64–67 (1997).
Li, J. et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 275, 1943–1947 (1997).
Steck, P. A. et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 15, 356–362 (1997).
Hollander, M. C., Blumenthal, G. M. & Dennis, P. A. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer 11, 289–301 (2011).
Shi, Y., Paluch, B. E., Wang, X. & Jiang, X. PTEN at a glance. J. Cell Sci. 125, 4687–4692 (2012).
Wang, X. & Jiang, X. Post-translational regulation of PTEN. Oncogene 27, 5454–5463 (2008).
Fata, J. E., Debnath, S., Jenkins, E. C. Jr. & Fournier, M. V. Nongenomic mechanisms of PTEN regulation. Int. J. Cell Biol. 2012, 379685 (2012).
Trotman, L. C. et al. Ubiquitination regulates PTEN nuclear import and tumour suppression. Cell 128, 141–156 (2007).
Wang, X. et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128, 129–139 (2007).
Maddika, S. et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–733 (2011).
Van Themsche, C., Leblanc, V., Parent, S. & Asselin, E. X-linked inhibitor of apoptosis protein (XIAP) regulates PTEN ubiquitination, content, and compartmentalization. J. Biol. Chem. 284, 20462–20466 (2009).
Ahmed, S.F. et al. The chaperone-assisted E3 ligase C terminus of Hsc70-interacting protein (CHIP) targets PTEN for proteasomal degradation. J. Biol. Chem. 287, 15996–16006 (2012).
Song, M. S. et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455, 813–817 (2008).
Wilkinson, K. D. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 11, 1245–1256 (1997).
Weigelt, B., Warne, P. H. & Downward, J. PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene 30, 3222–3233 (2011).
Hollestelle, A., Elstrodt, F., Nagel, J. H., Kallemeijn, W. W. & Schutte, M. Phosphatidylinositol-3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol. Cancer Res. 5, 195–201 (2007).
Zhao, X., Fiske, B., Kawakami, A., Li, J. & Fisher, D. E. Regulation of MITF stability by the USP13 deubiquitinase. Nat. Commun. 2, 414 (2011).
Song, M. S. et al. Nuclear PTEN regulates the APC-CDH1 tumour-suppressive complex in a phosphatase-independent manner. Cell 144, 187–199 (2011).
Lee, J. O. et al. Crystal structure of the PTEN tumour suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 99, 323–334 (1999).
Elstrom, R. L. et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 64, 3892–3899 (2004).
Manning, B. D. & Cantley, L. C. AKT/PKB signalling: navigating downstream. Cell 129, 1261–1274 (2007).
Warburg, O. On the origin of cancer cells. Science 123, 309–314 (1956).
Gustafson, S., Zbuk, K. M., Scacheri, C. & Eng, C. Cowden syndrome. Semin. Oncol. 34, 428–434 (2007).
Alimonti, A. et al. Subtle variations in Pten dose determine cancer susceptibility. Nat. Genet. 42, 454–458 (2010).
Perez-Tenorio, G. et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin. Cancer Res. 13, 3577–3584 (2007).
Chen, D. et al. LIFR is a breast cancer metastasis suppressor upstreamof the Hippo-YAP pathway and a prognostic marker. Nat. Med. 18, 1511–1517 (2012).
Yuan, J., Luo, K., Zhang, L., Cheville, J. C. & Lou, Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140, 384–396 (2010).
Giaccia, A. J. & Kastan, M. B. The complexity of p53 modulation: emerging patterns from divergent signals. Genes. Dev. 12, 2973–2983 (1998).
Salmena, L., Carracedo, A. & Pandolfi, P. P. Tenets of PTEN tumour suppression. Cell 133, 403–414 (2008).
Lee, C., Kim, J.S. & Waldman, T. PTEN gene targeting reveals a radiation-induced size checkpoint in human cancer cells. Cancer Res. 64, 6906–6914 (2004).
Kim, J. S., Lee, C., Bonifant, C. L., Ressom, H. & Waldman, T. Activation of p53-dependent growth suppression in human cells by mutations in PTEN or PIK3CA. Mol. Cell Biol. 27, 662–677 (2007).
Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. Mass spectrometricsequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996).
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Acknowledgements
We thank J. Yuan, Z. Gong, A. Sorokin, J. Wang, N. Li, L. Feng and L. Li for reagents and technical assistance. This work is supported by US National Institutes of Health grants R00CA138572 (to L.M.) and R01CA166051 (to L.M.) and a Cancer Prevention and Research Institute of Texas Scholar Award R1004 (to L.M.).
Author information
Authors and Affiliations
Contributions
J.Z. and L.M. conceived and designed the study and wrote the manuscript. J.Z. performed most of the experiments. P.Z. contributed to DUB library construction and in vitro deubiquitylation assays. Y.W. and M-C.H. performed studies on tissue microarrays of human patient samples. H-l.P. performed xenograft implantation. W.W. and J.C. assisted with tandem affinity purification and mass spectrometric analysis. S.M. provided the PTEN mutant constructs. M.W. assisted with animal care. D.C. assisted with lactate secretion assays. Y.S. maintained shRNA and ORF clones and assisted with glucose uptake assays.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Effects of five PTEN-interacting deubiquitinases on cell proliferation and colony formation.
(a) Five FLAG-tagged DUBs were expressed in MCF7 cells, immunoprecipitated with FLAG beads and immunoblotted with antibodies to PTEN and FLAG. (b) Growth curves of MCF7 cells transduced with USP7, USP8, USP10, USP13 or USP39. (c, d) Images (c) and quantification (d) of anchorage-independent growth of MCF7 cells transduced with USP7, USP8, USP10, USP13 or USP39. Data in (b) and (d) are the mean of 3 wells per group and error bars indicate s.e.m. The experiments were repeated 3 times. Statistical significance was determined by two-tailed, unpaired Student’s t test. Uncropped images of blots are shown in Supplementary Fig. 6.
Supplementary Figure 2 Regulation of PTEN and AKT signaling by USP13.
(a) Immunoblotting of USP13, PTEN and β-actin in a series of human breast cancer cell lines. (b) Immunoblotting of USP13, PTEN, p-AKT, AKT, p-FOXO1/3, FOXO1 and HSP90 in MDA-MB-231 cells transduced with USP13 alone or in combination with PTEN shRNA. (c) Immunoblotting of FLAG–USP13, HA-GFP and β-actin in 293T cells transfected with USP13 shRNA in combination with FLAG-tagged wild-type USP13 or an RNAi-resistant mutant of USP13 (USP13-RE). Co-transfected HA-GFP serves as the control for transfection. (d) Immunoblotting of USP13, PTEN, p-AKT, AKT, p-FOXO1/3, FOXO1 and HSP90 in USP13 shRNA-transduced SUM159 cells with or without ectopic expression of an RNAi-resistant mutant of USP13 (USP13-RE). (e) Immunoblotting of USP13, PTEN, p-AKT, AKT and β-actin in USP13 shRNA-transduced SUM159 cells with or without ectopic expression of PTEN. Cells were serum-starved and treated with 10 ng/ml insulin for 15 minutes. Uncropped images of blots are shown in Supplementary Fig. 6.
Supplementary Figure 3 USP13 does not alter PTEN mRNA levels.
(a) qPCR of PTEN and USP13 in USP13 shRNA-transduced SUM159 cells. (b) qPCR of PTEN in MDA-MB-231 cells transduced with wild-type USP13 or the USP13C345A mutant. Data in (a) and (b) are the mean of 3 triplicates per group and error bars indicate s.e.m. The experiments were repeated 3 times.
Supplementary Figure 4 USP7, USP10, USP8 and USP39 do not regulate PTEN protein levels.
Immunoblotting of the USP, PTEN and HSP90 in SUM159 cells with knockdown of USP7 (a), USP10 (b), USP8 (c) or USP39 (d). Uncropped images of blots are shown in Supplementary Fig. 6.
Supplementary Figure 5 Validation of the PTEN- and USP13-specific antibodies for immunohistochemistry.
The PTEN antibody was validated using BT549 (PTEN-null) and MCF7 (PTEN-positive) cell lines. The USP13 antibody was validated using parental SUM159 cells (USP13-positive) and USP13 shRNA-transduced SUM159 cells (USP13-depleted). To prepare sections, we fixed the cell pellet in formalin and embedded it in paraffin. Brown staining indicates positive immunoreactivity. Scale bar: 50 μm.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2937 kb)
Supplementary Table 1
Supplementary Information (XLSX 78 kb)
Supplementary Table 2
Supplementary Information (XLSX 9 kb)
Supplementary Table 3
Supplementary Information (XLSX 27 kb)
Rights and permissions
About this article
Cite this article
Zhang, J., Zhang, P., Wei, Y. et al. Deubiquitylation and stabilization of PTEN by USP13. Nat Cell Biol 15, 1486–1494 (2013). https://doi.org/10.1038/ncb2874
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2874
This article is cited by
-
USP43 stabilizes c-Myc to promote glycolysis and metastasis in bladder cancer
Cell Death & Disease (2024)
-
Exosomal USP13 derived from microvascular endothelial cells regulates immune microenvironment and improves functional recovery after spinal cord injury by stabilizing IκBα
Cell & Bioscience (2023)
-
USP13 drives lung squamous cell carcinoma by switching lung club cell lineage plasticity
Molecular Cancer (2023)
-
N6-methyladenosine-modified USP13 induces pro-survival autophagy and imatinib resistance via regulating the stabilization of autophagy-related protein 5 in gastrointestinal stromal tumors
Cell Death & Differentiation (2023)
-
Deubiquitinases in cancer
Nature Reviews Cancer (2023)