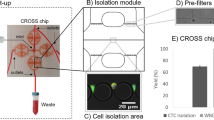
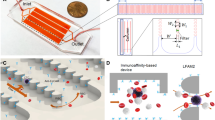
Abstract
The ability to isolate and analyze rare circulating tumor cells (CTCs) has the potential to further our understanding of cancer metastasis and enhance the care of cancer patients. In this protocol, we describe the procedure for isolating rare CTCs from blood samples by using tumor antigen–independent microfluidic CTC-iChip technology. The CTC-iChip uses deterministic lateral displacement, inertial focusing and magnetophoresis to sort up to 107 cells/s. By using two-stage magnetophoresis and depletion antibodies against leukocytes, we achieve 3.8-log depletion of white blood cells and a 97% yield of rare cells with a sample processing rate of 8 ml of whole blood/h. The CTC-iChip is compatible with standard cytopathological and RNA-based characterization methods. This protocol describes device production, assembly, blood sample preparation, system setup and the CTC isolation process. Sorting 8 ml of blood sample requires 2 h including setup time, and chip production requires 2–5 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
References
Lustberg, M., Jatana, K.R., Zborowski, M. & Chalmers, J.J. Emerging technologies for CTC detection based on depletion of normal cells. Recent Results Cancer Res. 195, 97–110 (2012).
Pantel, K., Brakenhoff, R.H. & Brandt, B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Med. 8, 329–340 (2008).
Yu, M., Stott, S., Toner, M., Maheswaran, S. & Haber, D.A. Circulating tumor cells: approaches to isolation and characterization. J. Cell Biol. 192, 373–382 (2011).
Maheswaran, S. et al. Detection of mutations in EGFR in circulating lung-cancer cells. N. Engl. J. Med. 359, 366–377 (2008).
Stott, S.L. et al. Isolation and characterization of circulating tumor cells from patients with localized and metastatic prostate cancer. Sci. Transl. Med. 2, 25ra23 (2010).
Yu, M. et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339, 580–584 (2013).
Yu, M. et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature 487, 510–513 (2012).
Zhang, L. et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Science Transl. Med. 5, 180ra48 (2013).
Baccelli, I. et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 31, 539–544 (2013).
Ramsköld, D. et al. Full-length mRNA-seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012).
Heitzer, E. et al. Complex tumor genomes inferred from single circulating tumor cells by array-CGH and next-generation sequencing. Cancer Res. 73, 2965–2975 (2013).
Sakaizawa, K. et al. Mutation analysis of BRAF and KIT in circulating melanoma cells at the single-cell level. Br. J. Cancer 106, 939–946 (2012).
Balasubramanian, P. et al. Multiparameter analysis, including EMT markers, on negatively enriched blood samples from patients with squamous cell carcinoma of the head and neck. PLoS ONE 7, e42048 (2012).
Payne, R.E. et al. Viable circulating tumour cell detection using multiplex RNA in situ hybridisation predicts progression-free survival in metastatic breast cancer patients. Br. J. Cancer 106, 1790–1797 (2012).
Haber, D.A., Gray, N.S. & Baselga, J. The evolving war on cancer. Cell Res. 145, 19–24 (2011).
Issadore, D. et al. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Sci. Transl. Med. 4, 141ra92 (2012).
Krivacic, R.T. et al. A rare-cell detector for cancer. Proc. Natl. Acad. Sci. USA 101, 10501–10504 (2004).
Hsieh, H.B. et al. High-speed detection of circulating tumor cells. Biosens. Bioelectron 21, 1893–1899 (2006).
Zhao, M. et al. An automated high-throughput counting method for screening circulating tumor cells in peripheral blood. Anal. Chem. 85, 2465–2471 (2013).
Ghossein, R.A. et al. Detection of circulating tumor cells in patients with localized and metastatic prostatic carcinoma: clinical implications. J. Clin. Oncol. 13, 1195–1200 (1995).
Simpson, S.J. et al. Detection of tumor cells in the bone marrow, peripheral blood, and apheresis products of breast cancer patients using flow cytometry. Exp. Hematol. 23, 1062–1068 (1995).
Witzig, T.E. et al. Detection of circulating cytokeratin-positive cells in the blood of breast cancer patients using immunomagnetic enrichment and digital microscopy. Clin. Cancer Res. 8, 1085–1091 (2002).
Deng, G. et al. Enrichment with anti-cytokeratin alone or combined with anti-EpCAM antibodies significantly increases the sensitivity for circulating tumor cell detection in metastatic breast cancer patients. Breast Cancer Res. 10, R69 (2008).
Talasaz, A.H. et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc. Natl. Acad. Sci. USA 106, 3970–3975 (2009).
Riethdorf, S. et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin. Cancer Res. 13, 920–928 (2007).
Lara, O., Tong, X., Zborowski, M. & Chalmers, J.J. Enrichment of rare cancer cells through depletion of normal cells using density and flow-through, immunomagnetic cell separation. Exp. Hematol. 32, 891–904 (2004).
Zheng, S. et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J. Chromatogr. A 1162, 154–161 (2007).
Vona, G. et al. Isolation by size of epithelial tumor cells. Am. J. Pathol. 156, 57–63 (2000).
Seal, S.H. A sieve for the isolation of cancer cells and other large cells from the blood. Cancer 17, 637–642 (1964).
Hou, H.W. et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci. Rep. 3, 1259 (2013).
Mohamed, H., Murray, M., Turner, J.N. & Caggana, M. Isolation of tumor cells using size and deformation. J. Chromatogr. A 1216, 8289–8295 (2009).
Hur, S.C., Henderson-MacLennan, N.K., McCabe, E.R.B. & Di Carlo, D. Deformability-based cell classification and enrichment using inertial microfluidics. Lab. Chip. 11, 912–920 (2011).
Gertler, R. et al. Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. Recent Results Cancer Res. 162, 149–155 (2003).
Litvinov, S.V., Velders, M.P., Bakker, H.A., Fleuren, G.J. & Warnaar, S.O. Ep-CAM: a human epithelial antigen is a homophilic cell-cell adhesion molecule. J. Cell Biol. 125, 437–446 (1994).
Herlyn, M., Steplewski, Z., Herlyn, D. & Koprowski, H. Colorectal carcinoma-specific antigen: detection by means of monoclonal antibodies. Proc. Natl. Acad. Sci. USA 76, 1438–1442 (1979).
Allard, W.J. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 10, 6897–6904 (2004).
de Bono, J.S. et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 14, 6302–6309 (2008).
Nagrath, S. et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450, 1235–1239 (2007).
Stott, S.L. et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl. Acad. Sci. USA 107, 18392–18397 (2010).
Miyamoto, D.T. et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2, 995–1003 (2012).
Thiery, J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454 (2002).
Kalluri, R. & Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420 (2009).
Münz, M. et al. Side-by-side analysis of five clinically tested anti-EpCAM monoclonal antibodies. Cancer Cell Int. 10, 44 (2010).
Pecot, C.V. et al. A novel platform for detection of CK+ and CK− CTCs. Cancer Discov. 1, 580–586 (2011).
Iinuma, H. et al. Detection of tumor cells in blood using CD45 magnetic cell separation followed by nested mutant allele-specific amplification of p53 and K-ras genes in patients with colorectal cancer. Int. J. Cancer 89, 337–344 (2000).
Bilkenroth, U. et al. Detection and enrichment of disseminated renal carcinoma cells from peripheral blood by immunomagnetic cell separation. Int. J. Cancer 92, 577–582 (2001).
Zigeuner, R.E., Riesenberg, R., Pohla, H., Hofstetter, A. & Oberneder, R. Immunomagnetic cell enrichment detects more disseminated cancer cells than immunocytochemistry in vitro. J. Urol. 164, 1834–1837 (2000).
Partridge, M., Phillips, E., Francis, R. & Li, S.R. Immunomagnetic separation for enrichment and sensitive detection of disseminated tumour cells in patients with head and neck SCC. J. Pathol. 189, 368–377 (1999).
Tong, X., Yang, L., Lang, J.C., Zborowski, M. & Chalmers, J.J. Application of immunomagnetic cell enrichment in combination with RT-PCR for the detection of rare circulating head and neck tumor cells in human peripheral blood. Cytometry 72B, 310–323 (2007).
Tkaczuk, K.H.R. et al. The significance of circulating epithelial cells in breast cancer patients by a novel negative selection method. Breast Cancer Res. Treat. 111, 355–364 (2007).
Liu, Z. et al. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J. Transl. Med. 9, 70 (2011).
Martin, V.M. et al. Immunomagnetic enrichment of disseminated epithelial tumor cells from peripheral blood by MACS. Exp. Hematol. 26, 252–264 (1998).
Miltenyi, S., Müller, W., Weichel, W. & Radbruch, A. High gradient magnetic cell separation with MACS. Cytometry 11, 231–238 (1990).
Zhang, H., Williams, P.S., Zborowski, M. & Chalmers, J.J. Binding affinities/avidities of antibody–antigen interactions: Quantification and scale-up implications. Biotechnol. Bioeng. 95, 812–829 (2006).
Lansdorp, P.M., Aalberse, R.C., Bos, R., Schutter, W.G. & Van Bruggen, E.F.J. Cyclic tetramolecular complexes of monoclonal antibodies: a new type of cross-linking reagent. Eur. J. Immunol. 16, 679–683 (1986).
Thomas, T.E., Sutherland, H.J. & Lansdorp, P.M. Specific binding and release of cells from beads using cleavable tetrameric antibody complexes. J. Immunol. Methods 120, 221–231 (1989).
Yang, L. et al. Optimization of an enrichment process for circulating tumor cells from the blood of head and neck cancer patients through depletion of normal cells. Biotechnol. Bioeng. 102, 521–534 (2009).
Ozkumur, E. et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci. Transl. Med. 5, 179ra47– (2013).
Huang, L.R., Cox, E.C., Austin, R.H. & Sturm, J.C. Continuous particle separation through deterministic lateral displacement. Science 304, 987–990 (2004).
Di Carlo, D., Irimia, D., Tompkins, R.G. & Toner, M. Continuous inertial focusing, ordering, and separation of particles in microchannels. Proc. Natl. Acad. Sci. USA 104, 18892–18897 (2007).
Pamme, N. & Wilhelm, C. Continuous sorting of magnetic cells via on-chip free-flow magnetophoresis. Lab Chip 6, 974–980 (2006).
Berger, M., Castelino, J., Huang, R., Shah, M. & Austin, R.H. Design of a microfabricated magnetic cell separator. Electrophoresis 22, 3883–3892 (2001).
Inglis, D.W., Riehn, R., Austin, R.H. & Sturm, J.C. Continuous microfluidic immunomagnetic cell separation. Appl. Phys. Lett. 85, 5093–5095 (2004).
Davis, J.A. et al. Deterministic hydrodynamics: taking blood apart. Proc. Natl. Acad. Sci. USA 103, 14779–14784 (2006).
Inglis, D.W., Davis, J.A., Austin, R.H. & Sturm, J.C. Critical particle size for fractionation by deterministic lateral displacement. Lab Chip 6, 655–658 (2006).
Loutherback, K. et al. Improved performance of deterministic lateral displacement arrays with triangular posts. Microfluid. Nanofluid. 9, 1143–1149 (2010).
Martel, J.M. & Toner, M. Inertial focusing dynamics in spiral microchannels. Phys. Fluids 24, 032001–032013 (2012).
Madou, M.J. Fundamentals of Microfabrication (CRC Press, 2002).
Boxshall, K. et al. Simple surface treatments to modify protein adsorption and cell attachment properties within a poly(dimethylsiloxane) micro-bioreactor. Surf. Interface Anal. 38, 198–201 (2006).
Yu, M et al. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev. 23, 1737–1742 (2009).
Acknowledgements
We express our gratitude to all patients and healthy volunteers who participated in this study and contributed blood samples. We thank L. Libby, O. Hurtado and A.J. Aranyosi for coordination of the research laboratories; D.M. Lewis and B. Hamza for expertise in device fabrication; and L. Nieman and J. Walsh for expertise in microscopy. This work was supported by the US National Institutes of Health (NIH) P41 Resource Center (M.T.); a NIH National Institute of Biomedical Imaging and Bioengineering Quantum Grant (M.T. and D.A.H.); Stand Up to Cancer (D.A.H., M.T. and S.M.); the Howard Hughes Medical Institute (D.A.H.); the Prostate Cancer Foundation and the Charles Evans Foundation (D.A.H. and M.T.); and Johnson and Johnson (M.T. and S.M.).
Author information
Authors and Affiliations
Contributions
All authors contributed to design of the CTC-iChip. N.M.K., P.S.S., F.F., V.P., E.O., N.K., K.S. and M.T. prepared the manuscript. E.J.L., J.M.M., P.C., J.Y., H.H., B.M., J.T., T.A.B., S.L.S., S.M., R.K. and D.A.H. commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Spintrap design and characterization.
(a, b) We designed and custom-made a closed centrifugation system, called Spintrap, for large volume, high throughput, and thin-layer preparation of CTCs. Due to its low fluidic resistance, the design allowed inline use with CTC-iChip with a processing time of 15 minutes for up to a fluid volume of 23 mL. The Spintrap consistently prevented air-drying and thus conserved native cell morphology. Using the Spintrap protocol (c, steps 88-92), and LBX1 cell line with cytoplasmic GFP label, we characterized cell attachment strength, yield, and resulting cellular morphology. (d) We performed automated fluorescence microscopy for quantifying efficiency of immobilization, and found the cell yield to be >95% at optimized conditions. (e, f) Confocal and epifluorescence microscopy revealed native cellular morphology at a relative centrifugal force of 50 g. A slight increase in cellular radius at 500 g and significant pancaking at 2,000 g was found, while cell circularity remained similar to a non-centrifuged control. We found centrifugation for 5 minutes at 50 g to be optimal for cell immobilization with high yield and without morphological disturbance. Cells collected with the Spintrap method showed diagnostic quality on cytological evaluation. Using Spintrap, we characterized CTCs from cancer patients isolated using CTC-iChip58.
Supplementary Figure 2 Spintrap fluidic simulation results.
Spintrap fluidic simulations show gradual deceleration and soft landing of cells to glass surface under 50g gravitational field.
Supplementary Figure 3 Calculated fluidic streamlines in CTC-iChip1.
a) Design of CTC-iChip1 posts used in fluidic simulations. b) 2D view of streamlines in the middle of the microfluidic channel, where the flow rate is highest. c) 3D view of streamlines.
Supplementary Figure 4 CTC-iChip1 assembly.
Detailed assembly of CTC-iChip1 with its manifold and tubing.
Supplementary Figure 5 Buffer and syringe cap schematics.
Schematic showing snorkel buffer cap and syringe cap. These parts help apply air pressure to blood in a syringe and buffer in a bottle using air-tight connections, and enable constant pressure operation of CTC-iChip
Supplementary information
Supplementary Figure 1
Spintrap design and characterization. (PDF 4349 kb)
Supplementary Figure 2
Spintrap fluidic simulation results. (PDF 1523 kb)
Supplementary Figure 3
Calculated fluidic streamlines in CTC-iChip1. (PDF 3409 kb)
Supplementary Figure 4
CTC-iChip1 assembly. (PDF 2067 kb)
Supplementary Figure 5
Buffer and syringe cap schematics. (PDF 641 kb)
Numerical simulation of the movement of a 10μm-diameter deformable cell surrounded by fluid in the Spintrap system centrifuged at 50g.
The applied centrifugal acceleration of 50g (direction: left to right) moves the cell toward the right wall where the glass slide is located. The custom-built finite-element software package PAK [ M. Kojic, R. Slavkovic, M. Zivkovic, N. Grujovic and N. Filipovic, PAK Finite Element Program for solid and fluid mechanics, mass and heat transfer, coupled problems and biomechanics., Mech. Eng. Dept. University of Kragujevac and R&D Center for Bioengineering Kragujevac, Serbia (2010)] was modified to solve for the fluid velocity field as well as the deformations of the cell, during a constant 50g acceleration. The video shows the resulting fluid velocity field (color scale bar indicates speed in μm/s) and depicts the gradual decrease in the cell speed as it approaches the glass slide (see also Supplementary Figure 2). The resulting minimal cell deformation is in good agreement with the experimentally observed cell morphology for 50g (see Supplementary Figure 1e). (AVI 1952 kb)
High-speed video of Deterministic Lateral Displacement of a WBC in blood.
A moving WBC is tracked within CTC-iChip2. WBC is deflected due to DLD whereas RBCs are following fluid streamlines. By the 15th second, the WBC is almost completely separated from the RBC population. The video was captured at 10× magnification while the stage was moving to enable tracking of the WBC. The video is played at ∼200× slower speed than the particles are moving in the channel. (AVI 16779 kb)
High-speed video of magnetophoretically separated cells entering outlets of CTC-iChip2.
PC3-9 cells being enriched by deflection of WBCs. The video is played at ∼200× slower than real-time. Note that this video was taken from a slightly different design, which has the product outlet in the middle and magnetophoresis setup pulling the WBCs to the sidewall. (AVI 5177 kb)
CTC-iChip2 Assembly.
Assembly of CTC-iChip2 with its magnetic manifold and their alignment is demonstrated. (MOV 43125 kb)
Supplementary Data
CTC-iChip2 Design CAD. (ZIP 354 kb)
Rights and permissions
About this article
Cite this article
Karabacak, N., Spuhler, P., Fachin, F. et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc 9, 694–710 (2014). https://doi.org/10.1038/nprot.2014.044
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.044
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.