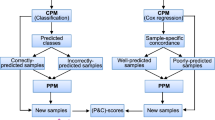
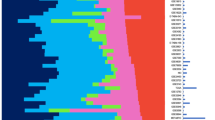
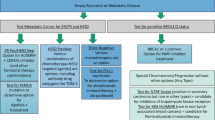
Abstract
Several gene-expression-based reference laboratory tests are now available for prognostication of patients diagnosed with breast cancer. For clinical oncologists, it is important to understand the clinical contexts for which these assays were developed in order to use them properly. This Review is aimed at providing a conceptual and technical overview of the steps involved in the development of gene-expression profiling-based prognostic assays. MammaPrint® and Oncotype DX®, two widely utilized assays, are compared with respect to differences in the clinical contexts for their development, technologies used, and clinical utilities with the aim of providing a guide to clinical oncologists for utilization of these assays.
Key Points
-
Clinical prognostic tests are most useful when developed and validated for a specific clinical context with a clear cut-off point for decision-making
-
There are two commercial reference laboratory tests based on gene-expression profiling (MammaPrint® and Oncotype DX®) that are either agency-approved or widely-accepted by the oncology community
-
These tests are not perfect and performances are similar
-
Their clinical utility should be determined based on specific clinical contexts
-
Oncotype DX® assay requires routinely processed formalin-fixed paraffin-embedded tumor tissue block, whereas the MammaPrint® assay requires snap-frozen tumor tissue or fresh tumor tissue procured in a special buffer
-
More studies are needed to examine the influence of intratumor heterogeneity or previous needle biopsy procedures on the performance of these assays
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Mook, S., van' t Veer, L. J., Rutgers, E. J., Piccart-Gebhart, M. J. & Cardoso, F. Individualization of therapy using Mammaprint: from development to the MINDACT Trial. Cancer Genomics Proteomics 4, 147–155 (2007).
Ross, J. S., Hatzis, C., Symmans, W. F., Pusztai, L. & Hortobágyi, G. N. Commercialized multigene predictors of clinical outcome for breast cancer. Oncologist 13, 477–493 (2008).
Kim, C., Taniyama, Y. & Paik, S. Gene expression-based prognostic and predictive markers for breast cancer: a primer for practicing pathologists. Arch. Pathol. Lab. Med. 133, 855–859 (2009).
Paik, S. et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351, 2817–2826 (2004).
van 't Veer, L. J. et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415, 530–536 (2002).
Loi, S. et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J. Clin. Oncol. 25, 1239–1246 (2007).
Ma, X. J. et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin. Cancer Res. 14, 2601–2608 (2008).
Liedtke, C. et al. Genomic grade index is associated with response to chemotherapy in patients with breast cancer. J. Clin. Oncol. 27, 3185–3191 (2009).
Albain, K. S. et al. Prognostic and predictive value of the 21-gene recurrence score assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol. 11, 55–65 (2010).
van de Vijver, M. J. et al. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 347, 1999–2009 (2002).
Wittner, B. S. et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin. Cancer Res. 14, 2988–2993 (2008).
Straver, M. E. et al. The 70-gene signature as a response predictor for neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. Treat. 119, 551–558 (2010).
Paik, S., Kim, C. Y., Song, Y. K. & Kim, W. S. Technology insight: application of molecular techniques to formalin-fixed paraffin-embedded tissues from breast cancer. Nat. Clin. Pract. Oncol. 2, 246–254 (2005).
Geiss, G. K. et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317–325 (2008).
Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 (1995).
Shi, L. et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 24, 1151–1161 (2006).
Masuda, N., Ohnishi, T., Kawamoto, S., Monden, M. & Okubo, K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 27, 4436–4443 (1999).
Cronin, M. et al. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am. J. Pathol. 164, 35–42 (2004).
Perou, C. M. et al. Molecular portraits of human breast tumours. Nature 406, 747–752 (2000).
Sørlie, T. et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. USA 98, 10869–10874 (2001).
Parker, J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 (2009).
Mook, S. et al. The 70-gene prognosis signature predicts early metastasis in breast cancer patients between 55 and 70 years of age. Ann. Oncol. 21, 717–722 (2010).
Straver, M. E. et al. The 70-gene signature as a response predictor for neoadjuvant chemotherapy in breast cancer. Breast Cancer Res. Treat. 119, 551–558 (2009).
Wittner, B. S. et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin. Cancer Res. 14, 2988–2993 (2008).
Glas, A. M. et al. Converting a breast cancer microarray signature into a high-throughput diagnostic test. BMC Genomics 7, 278 (2006).
Cardoso, F. et al. The MINDACT trial: the first prospective clinical validation of a genomic tool. Mol. Oncol. 1, 246–251 (2007).
Bogaerts, J. et al. Gene signature evaluation as a prognostic tool: challenges in the design of the MINDACT trial. Nat. Clin. Pract. Oncol. 3, 540–551 (2006).
Cardoso, F. et al. Clinical application of the 70-gene profile: the MINDACT trial. J. Clin. Oncol. 26, 729–735 (2008).
Paik, S. et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J. Clin. Oncol. 24, 3726–3734 (2006).
Sparano, J. A. TAILORx: trial assigning individualized options for treatment (Rx). Clin. Breast Cancer 7, 347–350 (2006).
Cronin, M. et al. Analytical validation of the Oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor-positive breast cancer. Clin. Chem. 53, 1084–1091 (2007).
Habel, L. A. et al. A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res. 8, R25 (2006).
Fan, C. et al. Concordance among gene-expression-based predictors for breast cancer. N. Engl. J. Med. 355, 560–569 (2006).
Schardt, J. A. et al. Genomic analysis of single cytokeratin-positive cells from bone marrow reveals early mutational events in breast cancer. Cancer Cell 8, 227–239 (2005).
Goldstein, L. J. et al. Prognostic utility of the 21-gene assay in hormone receptor-positive operable breast cancer compared with classical clinicopathologic features. J. Clin. Oncol. 26, 4063–4071 (2008).
Sotiriou, C. et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J. Natl Cancer Inst. 98, 262–272 (2006).
Toussaint, J. et al. Improvement of the clinical applicability of the Genomic Grade Index through a qRT-PCR test performed on frozen and formalin-fixed paraffin-embedded tissues. BMC Genomics 10, 424 (2009).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Dr. Soonmyung Paik declares he is a patent holder for the Oncotype DX® assay, produced by Genomic Health Inc. Dr. Paik is listed as one of the inventors for the Oncotype DX® assay but all rights have been assigned to the NSABP Foundation and Dr. Paik has no financial relationship with the company nor has he received any royalty based on the patent. C. Kim declares no competing interests.
Rights and permissions
About this article
Cite this article
Kim, C., Paik, S. Gene-expression-based prognostic assays for breast cancer. Nat Rev Clin Oncol 7, 340–347 (2010). https://doi.org/10.1038/nrclinonc.2010.61
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrclinonc.2010.61
This article is cited by
-
Association of serum cortisol and cortisone levels and risk of recurrence after endocrine treatment in breast cancer
Clinical and Experimental Medicine (2023)
-
Reproducibility and intratumoral heterogeneity of the PAM50 breast cancer assay
Breast Cancer Research and Treatment (2023)
-
Drug susceptibility testing of circulating lung cancer cells for personalized treatment
Medical Oncology (2022)
-
Prognostic gene expression signatures of breast cancer are lacking a sensible biological meaning
Scientific Reports (2021)
-
Advances in liquid biopsy using circulating tumor cells and circulating cell-free tumor DNA for detection and monitoring of breast cancer
Clinical and Experimental Medicine (2019)