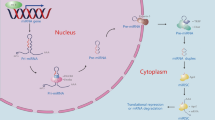
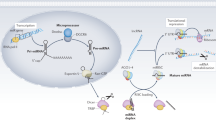
Abstract
Small, noncoding microRNAs (miRNAs) regulate diverse biological functions in the liver and increasing evidence suggests that they have a role in liver pathology. This Review summarizes advances in the field of miRNAs in liver diseases, inflammation and cirrhosis. MicroRNA-122, the most abundant miRNA in hepatocytes, has well-defined roles in HCV replication, and data indicate that it also serves as a viable therapeutic target. The role of miR-122 is also emerging in other liver diseases. Ample evidence exists for the important regulatory potential of other miRNAs in conditions associated with liver inflammation related to alcohol use, the metabolic syndrome or autoimmune processes. In addition, a broad array of miRNAs have been associated with the development of liver fibrosis both in animal models and human studies. The significance of the function and cellular distribution of miRNAs in the liver and the potential of miRNAs as a means of communication between cells and organs is discussed as well as the emerging utility of circulating miRNAs as biomarkers of different forms of liver damage and as early markers of disease and progression in hepatocellular carcinoma. Importantly, miRNA modulation in the liver represents a new therapeutic approach in the treatment armamentarium of hepatologists in the future.
Key Points
-
MicroRNAs (miRNAs) are encoded by genes and exert their intracellular effects by targeting post-transcriptional events on target genes
-
MiRNAs fine-tune all physiological and many pathological processes that are fundamental to normal liver functions and liver disease
-
The distribution and function of some miRNAs is cell-specific, and hepatocytes have the highest abundance of microRNA-122 expression in the body
-
In the liver, miRNAs have been shown to regulate processes such as inflammation, fibrosis, and lipid and glucose metabolism
-
Dysregulation in miRNAs is associated with liver diseases, such as hepatocellular carcinoma, viral hepatitis, alcoholic and nonalcoholic steatohepatitis and drug-induced liver injury
-
Extracellular miRNAs could serve as biomarkers of liver disease, but specificity might be a limitation
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355 (2004).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Sun, G. et al. SNPs in human miRNA genes affect biogenesis and function. RNA 15, 1640–1651 (2009).
Schoof, C. R., Botelho, E. L., Izzotti, A. & Vasques Ldos, R. MicroRNAs in cancer treatment and prognosis. Am. J. Cancer. Res. 2, 414–433 (2012).
Rottiers, V. & Naar, A. M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 13, 239–250 (2012).
Krutzfeldt, J. et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature 438, 685–689 (2005).
Esau, C. et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98 (2006).
Lanford, R. E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 (2010).
Cheung, O. et al. Nonalcoholic steatohepatitis is associated with altered hepatic microRNA expression. Hepatology 48, 1810–1820 (2008).
Wang, B. et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology 50, 1152–1161 (2009).
Bala, S. et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced and inflammatory liver diseases. Hepatology 56, 1946–1957 (2012).
Tsai, W. C. et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 122, 2884–2897 (2012).
Hsu, S. H. et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Invest. 122, 2871–2883 (2012).
Iliopoulos, D., Drosatos, K., Hiyama, Y., Goldberg, I. J. & Zannis, V. I. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J. Lipid Res. 51, 1513–1523 (2010).
Lee, J. et al. A pathway involving farnesoid X receptor and small heterodimer partner positively regulates hepatic sirtuin 1 levels via microRNA-34a inhibition. J. Biol. Chem. 285, 12604–12611 (2010).
Lee, J. & Kemper, J. K. Controlling SIRT1 expression by microRNAs in health and metabolic disease. Aging (Albany NY) 2, 527–534 (2010).
Meng, F. et al. Epigenetic regulation of miR-34a expression in alcoholic liver injury. Am. J. Pathol. 181, 804–817 (2012).
Vickers, K. C. et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology 57, 533–542 (2012).
O'Neill, L. A., Sheedy, F. J. & McCoy, C. E. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 11, 163–175 (2011).
Bala, S. & Szabo, G. MicroRNA signature in alcoholic liver disease. Int. J. Hepatol. http://dx.doi.org/10.1155/2012/498232.
Thai, T. H. et al. Regulation of the germinal center response by microRNA-155. Science 316, 604–608 (2007).
Bala, S. et al. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J. Biol. Chem. 286, 1436–1444 (2011).
Bala, S. et al. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J. Transl. Med. 10, 151 (2012).
Sidorkiewicz, M. et al. Expression of microRNA-155 precursor in peripheral blood mononuclear cells from Hepatitis C patients after antiviral treatment. Acta Virol. 54, 75–78 (2010).
Grek, M. et al. Coordinated increase of miRNA-155 and miRNA-196b expression correlates with the detection of the antigenomic strand of hepatitis C virus in peripheral blood mononuclear cells. Int. J. Mol. Med. 28, 875–880 (2011).
Zhang, Y. et al. Hepatitis C Virus-induced upregulation of miR-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology 56, 1631–1640 (2012).
Zhao, J. L. et al. NF-κB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl Acad. Sci. USA 108, 9184–9189 (2011).
Shaked, I. et al. MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity 31, 965–973 (2009).
Lagos, D. et al. miR-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat. Cell Biol. 12, 513–519 (2010).
Strum, J. C. et al. MicroRNA 132 regulates nutritional stress-induced chemokine production through repression of SirT1. Mol. Endocrinol. 23, 1876–1884 (2009).
Bihrer, V. et al. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am. J. Gastroenterol. 106, 1663–1669 (2011).
An, F. et al. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis 17, 702–716 (2012).
Yu, D. S. et al. The regulatory role of microRNA-1187 in TNF-α-mediated hepatocyte apoptosis in acute liver failure. Int. J. Mol. Med. 29, 663–668 (2012).
Sharma, A. D. et al. MicroRNA-221 regulates FAS-induced fulminant liver failure. Hepatology 53, 1651–1661 (2011).
Song, G. et al. MicroRNAs control hepatocyte proliferation during liver regeneration. Hepatology 51, 1735–1743 (2010).
Ng, R., Song, G., Roll, G. R., Frandsen, N. M. & Willenbring, H. A microRNA-21 surge facilitates rapid cyclin D1 translation and cell cycle progression in mouse liver regeneration. J. Clin. Invest. 122, 1097–1108 (2012).
Zhou, J. et al. Down-regulation of microRNA-26a promotes mouse hepatocyte proliferation during liver regeneration. PLoS ONE 7, e33577 (2012).
Pan, C. et al. Down-regulation of MiR-127 facilitates hepatocyte proliferation during rat liver regeneration. PLoS ONE 7, e39151 (2012).
Yuan, Q. et al. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology 57, 299–310 (2013).
Kalluri, R. & Weinberg, R. A. The basics of epithelial–mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009).
Kong, W. et al. MicroRNA-155 is regulated by the transforming growth factor β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol. Cell. Biol. 28, 6773–6784 (2008).
Gregory, P. A. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 (2008).
Park, S. M., Gaur, A. B., Lengyel, E. & Peter, M. E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 22, 894–907 (2008).
Korpal, M. & Kang, Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 5, 115–119 (2008).
Murakami, Y. et al. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE 6, e16081 (2011).
Jung, C. J. et al. Human ESC self-renewal promoting microRNAs induce epithelial-mesenchymal transition in hepatocytes by controlling the PTEN and TGFβ tumor suppressor signaling pathways. Mol. Cancer. Res. 10, 979–991 (2012).
Noetel, A., Kwiecinski, M., Elfimova, N., Huang, J. & Odenthal, M. microRNA are central players in anti- and profibrotic gene regulation during liver fibrosis. Front. Physiol. 3, 49 (2012).
He, Y. et al. The potential of microRNAs in liver fibrosis. Cell Signal. 24, 2268–2272 (2012).
Roderburg, C. et al. Micro-RNA profiling in human serum reveals compartment-specific roles of miR-571 and miR-652 in liver cirrhosis. PLoS ONE 7, e32999 (2012).
Wang, B. et al. miR-181b promotes hepatic stellate cells proliferation by targeting p27 and is elevated in the serum of cirrhosis patients. Biochem. Biophys. Res. Commun. 421, 4–8 (2012).
Roderburg, C. et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology 53, 209–218 (2011).
Kwiecinski, M. et al. Expression of platelet-derived growth factor-C and insulin-like growth factor I in hepatic stellate cells is inhibited by miR-29. Lab. Invest. 92, 978–987 (2012).
Mannaerts, I. et al. Class II HDAC inhibition hampers hepatic stellate cell activation by induction of microRNA-29. PLoS ONE 8, e55786 (2013).
Hand, N. J. et al. MicroRNA profiling identifies miR-29 as a regulator of disease-associated pathways in experimental biliary atresia. J. Pediatr. Gastroenterol. Nutr. 54, 186–192 (2012).
Marquez, R. T. et al. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab. Invest. 90, 1727–1736 (2010).
Venugopal, S. K. et al. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G101–G106 (2010).
He, Y. et al. MicroRNA-146a modulates TGF-β1-induced hepatic stellate cell proliferation by targeting SMAD4. Cell. Signal. 24, 1923–1930 (2012).
Li, J. et al. miR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. J. Hepatol. 58, 522–528 (2013).
Das, S. et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 110, 1596–1603 (2012).
Kren, B. T. et al. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 6, 65–72 (2009).
Bian, Z. et al. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 20, 1076–1078 (2010).
Hwang, H. W., Wentzel, E. A. & Mendell, J. T. A hexanucleotide element directs microRNA nuclear import. Science 315, 97–100 (2007).
Kim, D. H., Saetrom, P., Snove, O. Jr, & Rossi, J. J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl Acad. Sci. USA 105, 16230–16235 (2008).
Hansen, T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422 (2011).
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M. & Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 309, 1577–1581 (2005).
US National Library of Medicine. ClinicalTrials.gov[online], (2013).
Roberts, A. P., Lewis, A. P. & Jopling, C. L. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 39, 7716–7729 (2011).
Jopling, C. Liver-specific microRNA-122: biogenesis and function. RNA Biol. 9, 137–142 (2012).
Conrad, K. D. et al. microRNA-122 dependent binding of Ago2 protein to hepatitis C virus RNA is associated with enhanced RNA stability and translation stimulation. PLoS ONE 8, e56272 (2013).
Hou, W., Bukong, T. N., Kodys, K. & Szabo, G. Alcohol facilitates HCV RNA replication via up-regulation of miR-122 expression and inhibition of cyclin G1 in human hepatoma cells. Alcohol. Clin. Exp. Res. 37, 599–608 (2012).
Bukong, T. N., Hou, W., Kodys, K. & Szabo, G. Ethanol facilitates HCV replication via upregulation of GW182 and HSP90 in human hepatoma cells. Hepatology 57, 70–80 (2012).
Ura, S. et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology 49, 1098–1112 (2009).
Qiu, L. et al. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem. Biophys. Res. Commun. 398, 771–777 (2010).
Sarma, N. J. et al. Hepatitis C virus mediated changes in miRNA-449a modulates inflammatory biomarker YKL40 through components of the NOTCH signaling pathway. PLoS ONE 7, e50826 (2012).
Wang, K. et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl Acad. Sci. USA 106, 4402–4407 (2009).
Yoshioka, W., Higashiyama, W. & Tohyama, C. Involvement of microRNAs in dioxin-induced liver damage in the mouse. Toxicol. Sci. 122, 457–465 (2011).
Yokoi, T. & Nakajima, M. microRNAs as mediators of drug toxicity. Annu. Rev. Pharmacol. Toxicol. 53, 377–400 (2013).
Mohri, T. et al. Human CYP2E1 is regulated by miR-378. Biochem. Pharmacol. 79, 1045–1052 (2010).
Dolganiuc, A. et al. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol. Clin. Exp. Res. 33, 1704–1710 (2009).
Wanet, A., Tacheny, A., Arnould, T. & Renard, P. miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucleic Acids Res. 40, 4742–4753 (2012).
Tang, Y. et al. Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol. Clin. Exp. Res. 32, 355–364 (2008).
Yin, H. et al. MicroRNA-217 promotes ethanol-induced fat accumulation in hepatocytes by down-regulating SIRT1. J. Biol. Chem. 287, 9817–9826 (2012).
Yeligar, S., Tsukamoto, H. & Kalra, V. K. Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1α and microrNA-199. J. Immunol. 183, 5232–5243 (2009).
Xie, J. et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods 9, 403–409 (2012).
Venook, A. P., Papandreou, C., Furuse, J. & de Guevara, L. L. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist 15 (Suppl. 4), 5–13 (2010).
Hou, J. et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell 19, 232–243 (2011).
Calin, G. A. et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl Acad. Sci. USA 101, 11755–11760 (2004).
Ding, J. et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat. Cell Biol. 12, 390–399 (2010).
Fornari, F. et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene 27, 5651–5661 (2008).
Pineau, P. et al. miR-221 overexpression contributes to liver tumorigenesis. Proc. Natl Acad. Sci. USA 107, 264–269 (2010).
Park, J. K. et al. miR-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 71, 7608–7616 (2011).
Xiong, Y. et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 51, 836–845 (2010).
Fang, J. H. et al. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology 54, 1729–1740 (2011).
Braconi, C. et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 30, 4750–4756 (2011).
Murakami, Y. et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene 25, 2537–2545 (2006).
Kota, J. et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137, 1005–1017 (2009).
Ji, J. et al. MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 361, 1437–1447 (2009).
Braconi, C., Henry, J. C., Kogure, T., Schmittgen, T. & Patel, T. The role of microRNAs in human liver cancers. Semin. Oncol. 38, 752–763 (2011).
Jiang, J. et al. Association of microRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin. Cancer Res. 14, 419–427 (2008).
Coulouarn, C., Factor, V. M., Andersen, J. B., Durkin, M. E. & Thorgeirsson, S. S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene 28, 3526–3536 (2009).
Padgett, K. A. et al. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J. Autoimmun. 32, 246–253 (2009).
Banales, J. M. et al. Up-regulation of microRNA 506 leads to decreased Cl-/HCO3- anion exchanger 2 expression in biliary epithelium of patients with primary biliary cirrhosis. Hepatology 56, 687–697 (2012).
Poupon, R. et al. Genetic factors of susceptibility and of severity in primary biliary cirrhosis. J. Hepatol. 49, 1038–1045 (2008).
Salas, J. T. et al. Ae2a, b-deficient mice develop antimitochondrial antibodies and other features resembling primary biliary cirrhosis. Gastroenterology 134, 1482–1493 (2008).
Qin, B., Huang, F., Liang, Y., Yang, Z. & Zhong, R. Analysis of altered microRNA expression profiles in peripheral blood mononuclear cells from patients with primary biliary cirrhosis. J. Gastroenterol. Hepatol. 28, 543–550 (2013).
Castoldi, M. et al. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J. Clin. Invest. 121, 1386–1396 (2011).
Weber, J. A. et al. The microRNA spectrum in 12 body fluids. Clin. Chem. 56, 1733–1741 (2010).
Mitchell, P. S. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA 105, 10513–10518 (2008).
Shigehara, K. et al. Real-time PCR-based analysis of the human bile microRNAome identifies miR-9 as a potential diagnostic biomarker for biliary tract cancer. PLoS ONE 6, e23584 (2011).
Arroyo, J. D. et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl Acad. Sci. USA 108, 5003–5008 (2011).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Zernecke, A. et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2, ra81 (2009).
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D. & Remaley, A. T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–433 (2011).
Gibbings, D. J., Ciaudo, C., Erhardt, M. & Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 11, 1143–1149 (2009).
Vickers, K. C. & Remaley, A. T. Lipid-based carriers of microRNAs and intercellular communication. Curr. Opin. Lipidol. 23, 91–97 (2012).
Zhang, L. et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 22, 107–126 (2012).
Cermelli, S., Ruggieri, A., Marrero, J. A., Ioannou, G. N. & Beretta, L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS ONE 6, e23937 (2011).
Ji, F. et al. Circulating microRNAs in hepatitis B virus-infected patients. J. Viral Hepat. 18, e242–e251 (2011).
Qu, K. Z., Zhang, K., Li, H., Afdhal, N. H. & Albitar, M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J. Clin. Gastroenterol. 45, 355–360 (2011).
Xu, J. et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 50, 136–142 (2011).
Gui, J. et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin. Sci. (Lond.) 120, 183–193 (2010).
Tomimaru, Y. et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J. Hepatol. 56, 167–175 (2011).
Starkey Lewis, P. J. et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 54, 1767–1776 (2011).
Tryndyak, V. P. et al. Plasma microRNAs are sensitive indicators of inter-strain differences in the severity of liver injury induced in mice by a choline- and folate-deficient diet. Toxicol. Appl. Pharmacol. 262, 52–59 (2012).
Zhang, Y. et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin. Chem. 56, 1830–1838 (2010).
Zahm, A. M., Hand, N. J., Boateng, L. A. & Friedman, J. R. Circulating microRNA is a biomarker of biliary atresia. J. Pediatr. Gastroenterol. Nutr. 55, 366–369 (2012).
Murakami, Y. et al. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS ONE 7, e48366 (2012).
Szabo, G., Sarnow, P. & Bala, S. MicroRNA silencing and the development of novel therapies for liver disease. J. Hepatol. 57, 462–466 (2012).
Janssen, H. L. et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. http://dx.doi.org/10.1056/NEJMoa1209026
Stenvang, J., Petri, A., Lindow, M., Obad, S. & Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 3, 1 (2012).
Liu, X. Q., Song, W. J., Sun, T. M., Zhang, P. Z. & Wang, J. Targeted delivery of antisense inhibitor of miRNA for antiangiogenesis therapy using cRGD-functionalized nanoparticles. Mol. Pharm. 8, 250–259 (2011).
Su, J., Baigude, H., McCarroll, J. & Rana, T. M. Silencing microRNA by interfering nanoparticles in mice. Nucleic Acids Res. 39, e38 (2011).
Anand, S. et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat. Med. 16, 909–914 (2010).
Barrey, E. et al. Pre-microRNA and mature microRNA in human mitochondria. PLoS ONE 6, e20220 (2011).
Bandiera, S. et al. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS ONE 6, e20746 (2011).
Pegtel, D. M. et al. Functional delivery of viral miRNAs via exosomes. Proc. Natl Acad. Sci. USA 107, 6328–6333 (2010).
Zhang, Y. et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell 39, 133–144 (2010).
Kosaka, N. et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452 (2010).
Hu, G. et al. Exosome-mediated shuttling of microRNA-29 regulates HIV Tat and morphine-mediated neuronal dysfunction. Cell Death Dis. 3, e381 (2012).
Xin, H. et al. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells 30, 1556–1564 (2012).
Acknowledgements
The work of the authors is supported by NIAAA grant RO1-AA020744 (to G. Szabo).
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Szabo, G., Bala, S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol 10, 542–552 (2013). https://doi.org/10.1038/nrgastro.2013.87
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2013.87
This article is cited by
-
Exosomal microRNAs as biomarkers for viral replication in tofacitinib-treated rheumatoid arthritis patients with hepatitis C
Scientific Reports (2024)
-
The miR-3074/BMP7 axis regulates TGF-β-caused activation of hepatic stellate cells in vitro and CCl4-caused murine liver fibrosis in vivo
Human Cell (2024)
-
The intersection between alcohol-related liver disease and nonalcoholic fatty liver disease
Nature Reviews Gastroenterology & Hepatology (2023)
-
Biomarkers of liver diseases
Molecular Biology Reports (2023)
-
Missing Causality and Heritability of Autoimmune Hepatitis
Digestive Diseases and Sciences (2023)