Key Points
-
This Review focuses on the transcriptional control of lineage decisions during T-cell development. In particular, current progress in our understanding of the transcription factor network that regulates CD4/CD8 lineage choice is highlighted.
-
Recent work indicates that the specification of positively selected thymocytes to either the CD4 or CD8 lineage is a reversible step, but that lineage commitment is the subsequent irreversible step that determines T-cell fate.
-
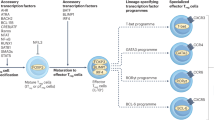
The transcription factors MYB, GATA-binding protein 3 (GATA3) and thymocyte selection-associated high-mobility group box (TOX) are lineage-specifying factors that cannot themselves enforce lineage choice, whereas RUNX (runt-related transcription factor) proteins and Th-POK (T-helper-inducing POZ/Kruppel-like factor) are lineage commitment factors that repress the development of the alternative fate.
-
Genetic studies have allowed the identification of enhancer and silencer elements in genes that are important for determining the fate of developing T cells, including Cd4, Cd8 and Zbtb7b (which encodes Th-POK). The binding of key transcription factors determines the activity of enhancer and silencer elements, thereby influencing gene expression and determining lineage choice. In this way, Th-POK has a role in commitment to the CD4 lineage, whereas RUNX3 is important for commitment to the CD8 lineage.
-
RUNX proteins also have a role in directing the differentiation of naive CD4+ T cells into different subsets of helper and regulatory T cells, and evidence suggests that the molecular mechanisms by which RUNX proteins regulate T-cell fate decisions are conserved between the thymus and periphery.
Abstract
Recent research has uncovered complex transcription factor networks that control the processes of T-cell development and differentiation. RUNX (runt-related transcription factor) proteins are among the many factors that have crucial roles in these networks. In this Review, we examine the mechanisms by which RUNX complexes act together with other transcription factors, such as Th-POK (T-helper-inducing POZ/Kruppel-like factor) and GATA-binding protein 3 (GATA3) in determining the CD4/CD8 lineage choice of developing thymocytes. In addition, we discuss evidence indicating that RUNX complexes are also involved in the differentiation of effector T-cell subsets and that the molecular mechanisms by which RUNX proteins regulate T-cell fate decisions are conserved between the thymus and periphery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Robey, E. A. et al. Thymic selection in CD8 transgenic mice supports an instructive model for commitment to a CD4 or CD8 lineage. Cell 64, 99–107 (1991).
Davis, C. B., Killeen, N., Crooks, M. E., Raulet, D. & Littman, D. R. Evidence for a stochastic mechanism in the differentiation of mature subsets of T lymphocytes. Cell 73, 237–247 (1993).
Yasutomo, K., Doyle, C., Miele, L., Fuchs, C. & Germain, R. N. The duration of antigen receptor signalling determines CD4+ versus CD8+ T-cell lineage fate. Nature 404, 506–510 (2000).
Singer, A. & Bosselut, R. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv. Immunol. 83, 91–131 (2004).
Brugnera, E. et al. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity 13, 59–71 (2000).
Singer, A., Adoro, S. & Park, J. H. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nature Rev. Immunol. 8, 788–801 (2008).
Ito, Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv. Cancer Res. 99, 33–76 (2008).
Speck, N. A. Core binding factor and its role in normal hematopoietic development. Curr. Opin. Hematol. 8, 192–196 (2001).
Okuda, T., van Deursen, J., Hiebert, S. W., Grosveld, G. & Downing, J. R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84, 321–330 (1996).
Ducy, P., Zhang, R., Geoffroy, V., Ridall, A. L. & Karsenty, G. Osf2/Cbfα1: a transcriptional activator of osteoblast differentiation. Cell 89, 747–754 (1997).
Komori, T. et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 (1997).
Li, Q. L. et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109, 113–124 (2002).
Levanon, D. et al. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 21, 3454–3463 (2002).
Inoue, K. et al. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nature Neurosci. 5, 946–954 (2002).
Sasaki, K. et al. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc. Natl Acad. Sci. USA 93, 12359–12363 (1996).
Wang, Q. et al. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell 87, 697–708 (1996).
Taniuchi, I. et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002). This study shows that RUNX complexes bind to the Cd4 silencer and are required for the silencing of CD4 expression.
Sawada, S., Scarborough, J. D., Killeen, N. & Littman, D. R. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell 77, 917–929 (1994).
Siu, G., Wurster, A. L., Duncan, D. D., Soliman, T. M. & Hedrick, S. M. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO J. 13, 3570–3579 (1994).
Leung, R. K. et al. Deletion of the CD4 silencer element supports a stochastic mechanism of thymocyte lineage commitment. Nature Immunol. 2, 1167–1173 (2001).
Zou, Y. R. et al. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nature Genet. 29, 332–336 (2001).
Taniuchi, I., Ellmeier, W. & Littman, D. R. The CD4/CD8 lineage choice: new insights into epigenetic regulation during T cell development. Adv. Immunol. 83, 55–89 (2004).
Taniuchi, I. & Littman, D. R. Epigenetic gene silencing by Runx proteins. Oncogene 23, 4341–4345 (2004).
Egawa, T., Tillman, R. E., Naoe, Y., Taniuchi, I. & Littman, D. R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204, 1945–1957 (2007).
Egawa, T. et al. Genetic evidence supporting selection of the Vα14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22, 705–716 (2005).
Sato, T. et al. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity 22, 317–328 (2005).
Ellmeier, W., Sawada, S. & Littman, D. R. The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu. Rev. Immunol. 17, 523–554 (1999).
Kioussis, D. & Ellmeier, W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nature Rev. Immunol. 2, 909–919 (2002).
Ellmeier, W., Sunshine, M. J., Losos, K. & Littman, D. R. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity 9, 485–496 (1998).
Hostert, A. et al. Hierarchical interactions of control elements determine CD8α gene expression in subsets of thymocytes and peripheral T cells. Immunity 9, 497–508 (1998).
Ellmeier, W., Sunshine, M. J., Losos, K., Hatam, F. & Littman, D. R. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity 7, 537–547 (1997).
Kohu, K. et al. Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single-positive lineage. J. Immunol. 174, 2627–2636 (2005).
Grueter, B. et al. Runx3 regulates integrin αE/CD103 and CD4 expression during development of CD4−/CD8+ T cells. J. Immunol. 175, 1694–1705 (2005).
He, X. et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature 433, 826–833 (2005).
Dave, V. P., Allman, D., Keefe, R., Hardy, R. R. & Kappes, D. J. HD mice: a novel mouse mutant with a specific defect in the generation of CD4+ T cells. Proc. Natl Acad. Sci. USA 95, 8187–8192 (1998).
Keefe, R., Dave, V., Allman, D., Wiest, D. & Kappes, D. J. Regulation of lineage commitment distinct from positive selection. Science 286, 1149–1153 (1999).
Bilic, I. & Ellmeier, W. The role of BTB domain-containing zinc finger proteins in T cell development and function. Immunol. Lett. 108, 1–9 (2007).
Klevit, R. E. Recognition of DNA by Cys2, His2 zinc fingers. Science 253, 1367, 1393 (1991).
Wang, L. et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4+ T cells. Nature Immunol. 9, 1122–1130 (2008).
Sun, G. et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nature Immunol. 6, 373–381 (2005). Along with reference 34, this study establishes the involvement of Th-POK in the differentiation of CD4+ T cells.
He, X. et al. CD4–CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity 28, 346–358 (2008).
Setoguchi, R. et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319, 822–825 (2008). References 41 and 42 identify a silencer element in the Zbtb7b locus that restricts full Th-POK expression to CD4-fated thymocytes.
Woolf, E. et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl Acad. Sci. USA 100, 7731–7736 (2003).
Muroi, S. et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nature Immunol. 9, 1113–1121 (2008).
Egawa, T. & Littman, D. R. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nature Immunol. 9, 1131–1139 (2008). Together with references 39 and 44, this study helped to define the hierarchy of transcription factors that are involved in T-cell lineage choice.
Wildt, K. F. et al. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J. Immunol. 179, 4405–4414 (2007).
Hernandez-Hoyos, G., Anderson, M. K., Wang, C., Rothenberg, E. V. & Alberola-Ila, J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity 19, 83–94 (2003).
Pai, S. Y. et al. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19, 863–875 (2003).
Maurice, D., Hooper, J., Lang, G. & Weston, K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 26, 3629–3640 (2007).
Wilkinson, B. et al. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nature Immunol. 3, 272–280 (2002).
Aliahmad, P. & Kaye, J. Development of all CD4 T lineages requires nuclear factor TOX. J. Exp. Med. 205, 245–256 (2008).
Ansel, K. M. et al. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nature Immunol. 5, 1251–1259 (2004).
Zheng, W. & Flavell, R. A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997).
Szabo, S. J. et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100, 655–669 (2000).
Djuretic, I. M. et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nature Immunol. 8, 145–153 (2007).
Naoe, Y. et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbfβ binding to the Il4 silencer. J. Exp. Med. 204, 1749–1755 (2007).
Ivanov, I. et al. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126, 1121–1133 (2006).
Fontenot, J. D., Gavin, M. A. & Rudensky, A. Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nature Immunol. 4, 330–336 (2003).
Zhang, F., Meng, G. & Strober, W. Interactions among the transcription factors Runx1, RORγt and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nature Immunol. 9, 1297–1306 (2008).
Zhou, L. et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature 453, 236–240 (2008).
Ichiyama, K. et al. Foxp3 inhibits RORγt-mediated IL-17A mRNA transcription through direct interaction with RORγt. J. Biol. Chem. 283, 17003–17008 (2008).
Yang, X. O. et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29, 44–56 (2008).
Ono, M. et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature 446, 685–689 (2007).
Ichikawa, M. et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nature Med. 10, 299–304 (2004).
Otto, F. et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771 (1997).
Niki, M. et al. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl Acad. Sci. USA 94, 5697–5702 (1997).
Acknowledgements
We thank our many colleagues who have contributed to the study of T-cell lineage choice and apologize for not including all references in this Review owing to space limitations. We are grateful to the members of our laboratories for helpful discussion. The work of I.T. was supported by grants from Precursory Research for Embryonic Science and Technology (PREST) and the Japan Science and Technology Agency (JST). The work of D.R.L. was supported by a grant from the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding authors
Related links
Glossary
- β-selection
-
A process that, through a cell-autonomous signalling cascade, leads to the proliferation, differentiation and survival of thymocytes that have successfully recombined the β-chain of the T-cell receptor (TCR) locus to express a functional pre-TCR on their cell surface.
- DNase hypersensitivity mapping
-
A technique that is used to investigate DNA regions that are likely to have regulatory roles in gene expression. It is based on the fact that the chromatin at regulatory regions is less condensed than at neighbouring regions, reflecting the need for increased accessibility to transcription factors, which results in higher sensitivity to digestion by endonucleases.
- ChIP-on-chip
-
A technique that combines chromatin immunoprecipitation, which determines the binding of transcription factors to specific regions of genomic DNA, with microarray chips to determine the binding of transcription factors in a genome-wide, unbiased manner.
- Hypomorphic mutant allele
-
A type of mutation in which either the altered gene product has a decreased level of activity or the wild-type gene product is expressed at a decreased level.
- CRE recombinase
-
A site-specific recombination system. Two short DNA sequences (loxP sites) are engineered to flank the target DNA. Expression of the recombinase CRE leads to excision of the intervening sequence. Depending on the type of promoter that controls CRE expression, CRE can be expressed at specific times during development or by specific subsets of cells.
- TH17 cell
-
A subset of CD4+ T helper (TH) cells that produce interleukin-17 (IL-17) and that are thought to be important in inflammatory and autoimmune diseases. Their generation involves IL-23 and IL-21, as well as the transcription factors retinoic-acid-receptor-related orphan receptor-γt and signal transducer and activator of transcription 3.
- Regulatory T (TReg) cell
-
A specialized type of CD4+ T cell that can suppress the responses of other T cells. TReg cells provide a crucial mechanism for the maintenance of peripheral self tolerance and are characterized by the expression of CD25 (also known as the α-chain of the interleukin-2 receptor) and the transcription factor forkhead box P3.
Rights and permissions
About this article
Cite this article
Collins, A., Littman, D. & Taniuchi, I. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat Rev Immunol 9, 106–115 (2009). https://doi.org/10.1038/nri2489
Issue Date:
DOI: https://doi.org/10.1038/nri2489
This article is cited by
-
Integrated single-cell chromatin and transcriptomic analyses of human scalp identify gene-regulatory programs and critical cell types for hair and skin diseases
Nature Genetics (2023)
-
TRANSPARENT: a Python tool for designing transcription factor regulatory networks
Soft Computing (2023)
-
RUNX1 inhibits the antiviral immune response against influenza A virus through attenuating type I interferon signaling
Virology Journal (2022)
-
The emerging role of transcription factor FOXP3 in thyroid cancer
Reviews in Endocrine and Metabolic Disorders (2022)
-
Selective deployment of transcription factor paralogs with submaximal strength facilitates gene regulation in the immune system
Nature Immunology (2019)