Abstract
During DNA replication, chromatin is reassembled by recycling of modified old histones and deposition of new ones. How histone dynamics integrates with DNA replication to maintain genome and epigenome information remains unclear. Here, we reveal how human MCM2, part of the replicative helicase, chaperones histones H3–H4. Our first structure shows an H3–H4 tetramer bound by two MCM2 histone-binding domains (HBDs), which hijack interaction sites used by nucleosomal DNA. Our second structure reveals MCM2 and ASF1 cochaperoning an H3–H4 dimer. Mutational analyses show that the MCM2 HBD is required for MCM2–7 histone-chaperone function and normal cell proliferation. Further, we show that MCM2 can chaperone both new and old canonical histones H3–H4 as well as H3.3 and CENPA variants. The unique histone-binding mode of MCM2 thus endows the replicative helicase with ideal properties for recycling histones genome wide during DNA replication.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Alabert, C. & Groth, A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 13, 153–167 (2012).
Hake, S.B. & Allis, C.D. Histone H3 variants and their potential role in indexing mammalian genomes: the “H3 barcode hypothesis”. Proc. Natl. Acad. Sci. USA 103, 6428–6435 (2006).
Margueron, R. & Reinberg, D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 11, 285–296 (2010).
Probst, A.V., Dunleavy, E. & Almouzni, G. Epigenetic inheritance during the cell cycle. Nat. Rev. Mol. Cell Biol. 10, 192–206 (2009).
Shibahara, K. & Stillman, B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 (1999).
Zhang, Z., Shibahara, K. & Stillman, B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408, 221–225 (2000).
Groth, A. et al. Regulation of replication fork progression through histone supply and demand. Science 318, 1928–1931 (2007).
Ishimi, Y., Komamura-Kohno, Y., Arai, K. & Masai, H. Biochemical activities associated with mouse Mcm2 protein. J. Biol. Chem. 276, 42744–42752 (2001).
Jasencakova, Z. et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell 37, 736–743 (2010).
Bochman, M.L. & Schwacha, A. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol. Mol. Biol. Rev. 73, 652–683 (2009).
Boos, D., Frigola, J. & Diffley, J.F. Activation of the replicative DNA helicase: breaking up is hard to do. Curr. Opin. Cell Biol. 24, 423–430 (2012).
McKnight, S.L. & Miller, O.L. Jr. Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell 12, 795–804 (1977).
Sogo, J.M., Stahl, H., Koller, T. & Knippers, R. Structure of replicating simian virus 40 minichromosomes: the replication fork, core histone segregation and terminal structures. J. Mol. Biol. 189, 189–204 (1986).
Annunziato, A.T. Split decision: what happens to nucleosomes during DNA replication? J. Biol. Chem. 280, 12065–12068 (2005).
Jackson, V. & Chalkley, R. A new method for the isolation of replicative chromatin: selective deposition of histone on both new and old DNA. Cell 23, 121–134 (1981).
Annunziato, A.T. Assembling chromatin: the long and winding road. Biochim. Biophys. Acta 1819, 196–210 (2013).
Smith, S. & Stillman, B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58, 15–25 (1989).
Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004).
Tyler, J.K. et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402, 555–560 (1999).
Burgess, R.J. & Zhang, Z. Histone chaperones in nucleosome assembly and human disease. Nat. Struct. Mol. Biol. 20, 14–22 (2013).
Ransom, M., Dennehey, B.K. & Tyler, J.K. Chaperoning histones during DNA replication and repair. Cell 140, 183–195 (2010).
Foltman, M. et al. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Reports 3, 892–904 (2013).
Ishimi, Y., Komamura, Y., You, Z. & Kimura, H. Biochemical function of mouse minichromosome maintenance 2 protein. J. Biol. Chem. 273, 8369–8375 (1998).
Richet, N. et al. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork. Nucleic Acids Res. 43, 1905–1917 (2015).
English, C.M., Adkins, M.W., Carson, J.J., Churchill, M.E. & Tyler, J.K. Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006).
Natsume, R. et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 446, 338–341 (2007).
Loyola, A., Bonaldi, T., Roche, D., Imhof, A. & Almouzni, G. PTMs on H3 variants before chromatin assembly potentiate their final epigenetic state. Mol. Cell 24, 309–316 (2006).
Mello, J.A. & Almouzni, G. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11, 136–141 (2001).
Groth, A. et al. Human Asf1 regulates the flow of S phase histones during replicational stress. Mol. Cell 17, 301–311 (2005).
Forsburg, S.L. Eukaryotic MCM proteins: beyond replication initiation. Microbiol. Mol. Biol. Rev. 68, 109–131 (2004).
Dimitrova, D.S., Todorov, I.T., Melendy, T. & Gilbert, D.M. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 146, 709–722 (1999).
Montagnoli, A. et al. Identification of Mcm2 phosphorylation sites by S-phase-regulating kinases. J. Biol. Chem. 281, 10281–10290 (2006).
Ge, X.Q., Jackson, D.A. & Blow, J.J. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 21, 3331–3341 (2007).
Gillespie, P.J. & Blow, J.J. Clusters, factories and domains: the complex structure of S-phase comes into focus. Cell Cycle 9, 3218–3226 (2010).
Alabert, C. et al. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 29, 585–590 (2015).
Bodor, D.L., Valente, L.P., Mata, J.F., Black, B.E. & Jansen, L.E. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol. Biol. Cell 24, 923–932 (2013).
Latreille, D., Bluy, L., Benkirane, M. & Kiernan, R.E. Identification of histone 3 variant 2 interacting factors. Nucleic Acids Res. 42, 3542–3550 (2014).
Tachiwana, H. et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476, 232–235 (2011).
Dunleavy, E.M. et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 137, 485–497 (2009).
Foltz, D.R. et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 137, 472–484 (2009).
Kaufman, P.D., Kobayashi, R. & Stillman, B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 11, 345–357 (1997).
Hoek, M. & Stillman, B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc. Natl. Acad. Sci. USA 100, 12183–12188 (2003).
Klapholz, B. et al. CAF-1 is required for efficient replication of euchromatic DNA in Drosophila larval endocycling cells. Chromosoma 118, 235–248 (2009).
Mejlvang, J. et al. New histone supply regulates replication fork speed and PCNA unloading. J. Cell Biol. 204, 29–43 (2014).
Houlard, M. et al. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2, e181 (2006).
Song, Y. et al. CAF-1 is essential for Drosophila development and involved in the maintenance of epigenetic memory. Dev. Biol. 311, 213–222 (2007).
Groth, A. Replicating chromatin: a tale of histones. Biochem. Cell Biol. 87, 51–63 (2009).
Xu, M. et al. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science 328, 94–98 (2010).
Huang, C. et al. H3.3–H4 tetramer splitting events feature cell-type specific enhancers. PLoS Genet. 9, e1003558 (2013).
Hu, H. et al. Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP. Genes Dev. 25, 901–906 (2011).
Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Amaro, A.C. et al. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat. Cell Biol. 12, 319–329 (2010).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 (2002).
Fujita, M., Kiyono, T., Hayashi, Y. & Ishibashi, M. In vivo interaction of human MCM heterohexameric complexes with chromatin: possible involvement of ATP. J. Biol. Chem. 272, 10928–10935 (1997).
Jansen, L.E., Black, B.E., Foltz, D.R. & Cleveland, D.W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 176, 795–805 (2007).
Acknowledgements
We thank the beamline staff at the synchrotrons at the Argonne National Laboratory (NE-CAT) and the Brookhaven National Laboratory (XL-29) for technical assistance. We thank H. Wu for access to SEC-MALS equipment for molecular-weight measurements. We thank Y. Feng and C. Alabert for help with immunofluorescence and PLA analysis. We thank C. Alabert for comments on the manuscript and Z. Jasencakova for help with experiments and for input in the manuscript and the model. We also thank P. Meraldi (University of Geneva) and L. Jansen (Gulbenkian Institute) for reagents and K. Labib (University of Dundee) for sharing information before publication. D.J.P. was supported in part by grants from the Leukemia and Lymphoma Society (LLS 7006-13) and the Starr foundation (I5-A554). A.G. is supported as a European Molecular Biology Organization Young Investigator, and her research is supported by the Danish National Research Foundation to the Center for Epigenetics (DNRF82), the European Commission ITN FP7 'aDDRess', a European Research Council Starting Grant (ERC2011StG, no. 281,765), the Danish Cancer Society, the Danish Medical Research Foundation and the Lundbeck Foundation.
Author information
Authors and Affiliations
Contributions
H.H. conceived and led the generation of cassettes for crystallization of the complexes, and H.H. solved the structures under the supervision of D.J.P. C.B.S. and A.G. conceived and led the functional studies. G.S. performed immunoprecipitation of HA–H3.1 from chromatin. M.H. analyzed histone incorporation by SNAP-tag assay. A.S. carried out lentiviral transduction and cell characterization. C.G.-A. helped with cloning and data analysis. S.C. performed SEC-MALS molecular-weight measurements. H.H., C.B.S., A.G. and D.J.P. wrote the manuscript, and all authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
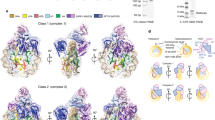
Supplementary Figure 1 Biochemical characterization of different MCM2 HBD–H3–H4 complexes and structural comparisons.
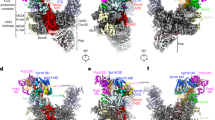
(a) His-pulldown of MCM2 HBD with recombinant histones. (b) The MCM2 HBD(61–130)–H3.3(Δ56) –H4 tetramer (complex 1) and MCM2 HBD(43–160)–H3.3(Δ56) –H4 tetramer (complex 2) were analyzed by gel-filtration assay. (c) SDS-PAGE analysis of the peak fractions from panel b. (d) The MCM2 HBD(43–160)–H3.2–H4 tetramer (complex 6), MCM2 HBD(43–160)–H3.2–H4 dimer–ASF1a(1-172) (complex 5) and H3.2–H4 tetramer were analyzed as in panel b. (e) SDS-PAGE analysis of the peak fractions from panel d. (f) SEC-MALS assay. The apparent Mw determined by SEC-MALS of MCM2 HBD–H3–H4 tetramer complex is 93.5 kDa (errors 3%, with the expected Mw about 80.0 kDa). The apparent Mw of MCM2 HBD–H3–H4 dimer–ASF1 complex is 64.3 kDa (errors 4%, with the expected Mw about 60.0 kDa). (g) A stereo view following superposition of the two halves of the MCM2 HBD–H3–H4 tetramer complex. One half was in color and the other half was in silver. (h) The reported structure (Richet, N. et al., Nucleic Acids Res 43, 1905-1917, 2015) observed a 1:1:1 MCM2 HBD–H3.2–H4 dimer complex in the asymmetric unit (AU) and a ‘2:2:2’ complex could be generated through crystallographic symmetric operation, while our structure directly observed a 2:2:2 MCM2 HBD–H3.3–H4 tetramer complex in the AU. Panel h showing the superposition of the generated ‘2:2:2’ structure (in silver) with our observed 2:2:2 structure (in color) with an rmsd of 1.3 Å. The major differences in conformations are pointed out with arrows. (i) A stereo view following superposition of the H3–H4 tetramers from the structures of MCM2 HBD–H3-H4 tetramer complex (in color) and nucleosome (in silver). The H2A docking domain from the nucleosome was also shown.
Supplementary Figure 2 Selective details of intermolecular contacts of our crystal structures and biochemical characterizations of different MCM2 HBD–H3–H4 –ASF1 complexes.
(a–c) Each panel highlighting the key interactions in the S1 (panel a), S2 (panel b) and S3 (panel c) regions of MCM2 HBD in the MCM2 HBD–H3–H4 tetramer complex. (d) GST-pulldown of ASF1 WT and V94R mutant with recombinant H3–H4 tetramers and prepurified MCM2 HBD–H3–H4 tetramer complex. (e) The prepurified MCM2 HBD(43–160)–H3–H4 tetramer (P1), ASF1b(1–158) (P3) and MCM2 HBD(43–163)–H3(EE) –H4 dimer (a constitutive H3–H4 dimer with dual mutation L126E I130E on H3) (P4) were analyzed by gel filtration and served as controls; the prepurified MCM2 HBD(43–160)–H3–H4 tetramer (20 nmole) was incubated with ASF1b(1–158) (12 nmole) and the reaction mixture was analyzed. Two peaks P1’ (not reacted complex) and P2 (reaction product) were expected and observed. (f) The peak fractions from panel e were analyzed on SDS-PAGE. (g) The MCM2 HBD(61–130)-linker-ASF1b(1–158)–H3.3(Δ56) –H4 dimer (complex 3) and MCM2 HBD(43-160)–H3.2(Δ55) –H4 dimer–ASF1a(1–172) (complex 4) were analyzed as in panel b of Supplementary Fig. 1. (h) SDS-PAGE analysis of the peak fractions from panel g. (i–k) Each panel highlighting the key interactions in the S1 (panel i), S2 (panel j) and S3 (panel k) regions of MCM2 HBD in the MCM2 HBD–H3–H4 dimer–ASF1 complex. For clarity, the ASF1 segment was omitted in panel k.
Supplementary Figure 3 Details of intermolecular contacts of ASF1–H3–H4 dimer complex and structural comparisons of different chaperone–H3–H4 complexes.
(a, b) Views of details of the intermolecular interactions between ASF1 and H3–H4 dimer from PDB 2IO5 (Natsume, R. et al., Nature 446, 338-341, 2007). Panel a highlighting the interactions of ASF1 with the helices α2 and α3 of H3, and panel b highlighting the interactions of ASF1 with the βc strand and C-terminus of H4. (c) Structural comparison of the ASF1–H3–H4 dimer complex (colored in silver) and our MCM2 HBD–H3–H4 dimer–ASF1 complex (in color). (d, e) Following the comparison shown in panel c, the RMSDs of residues in H3 (panel d) and H4 (panel e) were plotted respectively. The conformational differences on H3–H4 dimer caused upon binding of MCM2 HBD are spread out and include the L1 loop, the α2 helix and the L2 loop of H3 (panel d), as well as the α1 helix, the tip of α2 helix, the L2 loop and especially the α3 helix of H4 (panel e). (f) Structural comparison of one half of the MCM2 HBD–H3–H4 tetramer complex (colored in silver) with the MCM2 HBD–H3–H4 dimer–ASF1 complex (in color). (g, h) Following the comparison shown in panel f, the RMSDs of residues in H3 (panel g) and H4 (panel h) were plotted respectively.
Supplementary Figure 4 Structural comparison emphasizing the local differences and biochemical characterizations of the linker complex used for crystallization.
(a) Structural comparison as in panel c of Supplementary Fig. 3, emphasizing the local differences on H3 and H4. All the differences were seen along the MCM2 HBD binding interface on H3–H4. (b, c) Structural comparison as in panel f of Supplementary Fig. 3, emphasizing a 12o rotational difference in the α3 helix of H3 between conformations (panel b); and emphasizing the difference in the C-terminal tail of H4 between conformations, which undergoes a disorder to order transition (formation of a βc strand) with ASF1 (panel c). (d) Gel-filtration analysis of the complex with linker (complex 3) and without linker (complex 3’). The complex 3’ was similar to complex 3 except lacking the 12-mer covalent linker. (e) The peak fractions from panel d were analyzed on SDS-PAGE. (f) The tetrasome assembly assay. Lane 1-4, controls. Lane 5, excess H3–H4 tetramer mixed with DNA in physiological salt condition (150 mM NaCl) forming high aggregation. Lane 6, the prepurified complex of MCM2 HBD(43–160)–H3–H4 dimer–ASF1a(1–172) had tetrasome assembly activity. Lane 7, the prepurified complex of MCM2 HBD(61–130)-linker-ASF1b(1–158)–H3–H4 dimer had tetrasome assembly activity on linear Widom 601 DNA, though it shifted the balance among tetrasome and unassigned band (‘*’).
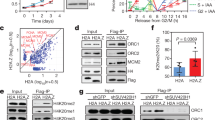
Supplementary Figure 5 Input controls for the immunoprecipitation reactions.
(a – f), Western blot of inputs for the immunoprecipitation reactions shown in Fig. 3a (panel a), Fig. 4a (panel b), Fig. 4c (panel c), Fig. 4e (panel d), Fig. 6a (panel e) and Fig. 6f (panel f).
Supplementary Figure 6 MCM2 binding is not required for incorporation of H3.1–H4 into chromatin but is important for stability of H3.1–H4.
(a) Representative images of newly synthesized TMR* labelled SNAP-HA-H3.1 wt and R63A K64A as described in Fig. 3b. Scale bar, 50 μm. (b) Analysis of total chromatin-bound and soluble SNAP-HA-H3.1 in stable cell lines used in Fig. 3b&c. Total SNAP-HA-H3.1 wt or R63A K64A were measured by TMR* labelling and direct fixation. The dot-plot showed median TMR* intensities from 3 independent experiments each including 5 technical replicates of total > 5,000 TMR* positive cells. Error bars, s.d. n=3. Unpaired t-test: *, P < 0.05. (c) SNAP-HA-H3.1 mRNA levels in stable cell lines used to measure the histone stability in Fig. 3c. Expression of e-H3 mRNA levels was measured by qPCR using two different primer sets, normalized to GAPDH and shown relative to wt. Error bars, s.d n=3 (for primer-set 2 wt, n=2). (d , e) Immunofluorescence of U-2-OS cells conditionally expressing Flag-HA-MCM2 WT or Y81A Y90A mutant. Cells were induced by tetracyclin (tet) for 24 hours and fixed directly (d) or pre-extracted (e) by CSK-T. HA staining confirmed induction of FLAG-HA-MCM2 in non-extracted cells (d) and showed typical cell cycle dependent MCM2 staining patterns in pre-extracted cells (e). Representative images of typical G1, early S and mid S phase were shown. Note: because MCM2-7 is unloaded as S phase progresses, late S and G2 cells are negative for MCM2. Flag-HA-MCM2 patterns were scored in 3 independent experiments (n=3), error-bars represent standard deviation, > 200 HA positive cells were scored per experiment. Scale bars, 20 μm (d) and 5 μm (e).
Supplementary Figure 7 ASF1a colocalizes with MCM2 and CDC45.
(a –c) PLA of endogenous chromatin-bound ASF1a with MCM2 (panels a, c) and CDC45 (panel b) as described in Fig. 6 b–d. Images showed a large field of pre-extracted cells to complement the single cells shown in Fig. 6. Scale bars were 16 μm. (d) Interactions of MCM2 HBD and H3.2–H4 in the MCM2 HBD–H3.2–H4dimer–ASF1 complex, highlighting the region S2 as in Fig. 1e. (e) Alignment of the MCM2 HBD binding region in histone H3 variants, showing strict conservation in red and conservation of properties in green. (f) Superposition of the H3–H4 tetramer from the MCM2 HBD–H3–H4 tetramer structure (in color) and the CENPA–H4 tetramer from the CENPA nucleosome structure (in silver).
Supplementary Figure 8 Biochemical characterizations of the MCM2–CENPA–H4 tetramer complex.
(a) Gel-filtration analyses of the CENPA–H4 tetramer and MCM2 HBD(43–160)–CENPA–H4 tetramer complex (complex 7) in high salt buffer (1 M NaCl). (b) The peak fractions from panel a were analyzed by SDS-PAGE. (c) The same analyses as in panel a, but with a low salt buffer (0.3 M NaCl). (d, e) IP of Flag-HA-MCM2 WT and its H3–H4-binding mutant from cells stably expressing EGFP-CENPA (Amaro, A.C. et al., Nat Cell Biol 12, 319-329, 2010).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Note (PDF 13463 kb)
Supplementary Data Set 1
Uncropped Images of gels (PDF 1335 kb)
Rights and permissions
About this article
Cite this article
Huang, H., Strømme, C., Saredi, G. et al. A unique binding mode enables MCM2 to chaperone histones H3–H4 at replication forks. Nat Struct Mol Biol 22, 618–626 (2015). https://doi.org/10.1038/nsmb.3055
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3055
This article is cited by
-
Structure of a histone hexamer bound by the chaperone domains of SPT16 and MCM2
Science China Life Sciences (2024)
-
The histone chaperone SPT2 regulates chromatin structure and function in Metazoa
Nature Structural & Molecular Biology (2024)
-
H4S47 O-GlcNAcylation regulates the activation of mammalian replication origins
Nature Structural & Molecular Biology (2023)
-
Symmetric inheritance of parental histones governs epigenome maintenance and embryonic stem cell identity
Nature Genetics (2023)
-
The patterns and participants of parental histone recycling during DNA replication in Saccharomyces cerevisiae
Science China Life Sciences (2023)