Abstract
The tumor suppressor gene p53 has been implicated in the regulation of epithelial–mesenchymal transition (EMT) and tumor metastasis by regulating microRNA (miRNA) expression. Here, we report that mutant p53 exerts oncogenic functions and promotes EMT in endometrial cancer (EC) by directly binding to the promoter of miR-130b (a negative regulator of ZEB1) and inhibiting its transcription. We transduced p53 mutants into p53-null EC cells, profiled the miRNA expression by miRNA microarray and identified miR-130b as a potential target of mutant p53. Ectopic expression of p53 mutants repressed the expression of miR-130b and triggered ZEB1-dependent EMT and cancer cell invasion. Loss of an endogenous p53 mutation increased the expression of miR-130b, which resulted in reduced ZEB1 expression and attenuation of the EMT phenotype. Furthermore, re-expression of miR-130b suppressed mutant p53-induced EMT and ZEB1 expression. Importantly, the expression of miR-130 was significantly reduced in EC tissues, and patients with higher expression levels of miR-130b survived longer. These data provide a novel understanding of the roles of p53 gain-of-function mutations in accelerating tumor progression and metastasis through modulation of the miR-130b–ZEB1 axis.
Similar content being viewed by others
Introduction
Epithelial–mesenchymal transition (EMT) is a transcriptional process that has a key role in regulating embryonic morphogenesis and cancer metastasis. During EMT, epithelial cells lose their polarization and homotypic cell adhesion, resulting in a more motile, spindle-like morphology with increased invasiveness.1 At the molecular level, EMT occurs as a result of the activity of several transcriptional factors, such as ZEB1/2, Twist, BMI-1, Snail, and Slug, which suppress expression of the epithelial marker E-cadherin and induce the mesenchymal genes N-cadherin and Vimentin.1 However, the mechanisms and pathways that drive EMT programs are not fully understood.
Non-coding microRNAs (miRNAs), including miR-200 and miR-194/192 family members, have been identified as negative regulators of EMT and metastasis by repressing the expression of ZEB1/2.2 The overexpression of miR-200 and let-7b in gemcitabine-resistant pancreatic cancer cells induces the mesenchymal–epithelial transition, which is the reverse process of EMT.3 Moreover, miR-194 is critical for maintaining the hepatic epithelial cell phenotype and inhibits metastasis by targeting several EMT activator genes.4
Recently, a regulatory connection between p53 signaling and miRNA-mediated EMT has been demonstrated. Wild-type (WT) p53 directly activates the transcription of miR-200c and miR-192 family members, which leads to ZEB1/2 downregulation and repression of EMT.5 Furthermore, mutation of p53 can promote EMT and the aggressive potential of tumor cells by inhibiting WT p53-miR-200c pathways through dominant-negative effects on WT p53.6 However, besides the dominant-negative effects upon WT p53, increasing evidence suggests that p53 mutations acquire additional oncogenic functions, such as a gain-of-function (GOF), which actively drive cells toward invasion and metastasis7 through transactivation or transrepression of a large set of genes involved in regulation of cell adhesion, migration and proliferation.8 In agreement with these findings, previous studies have found that overexpression of miRNAs (miR-181b and miR-200c) is associated with either p53 mutations or shorter patient survival in human colon cancer,9 indicating that mutant p53 may exert GOF activities and promote EMT by modulating miRNAs.
Here, we identified a novel mechanism by which mutant p53 demonstrates GOF effects to facilitate EMT and cancer cell invasion by repressing miR-130b, an inhibitor of ZEB1. We further demonstrated that the expression of miR-130 was significantly reduced in endometrial cancer (EC) tissues, and patients with higher expression levels of miR-130b survived longer. Thus, these data suggest that restoration of miR-130b may have therapeutic value in tumors expressing mutant p53.
Results
Mutant p53 GOF contributes to EMT in EC cells
Although mutant p53 GOF has been shown to promote EMT by upregulating Twist in prostate cancer cells,10 to date, the role of mutant p53 GOF in initiating EMT during EC progression remains unknown. To explore this issue, we used p53-null HEC-50 cells11 stably transduced with vectors encoding p53 mutations R273H, R175H or C135Y, as well as an empty vector. Polyclonal cell lines were generated to omit clonal variation. The p53 protein level was verified using the anti-p53 (DO-7) antibody, which recognizes both WT and mutant p53.
Enforced expression of these p53 mutants induce a shift in cell morphology from a paved stone epithelial appearance to more mesenchymal phenotypes, with loss of cell-to-cell contact and increased cell spreading (Figure 1a). These morphological changes were accompanied by the upregulation of mesenchymal genes, including Twist, ZEB1, BMI-1, Snail, N-cadherin and Vimentin, and decreased expression of the epithelial marker E-cadherin (Figure 1b). To examine whether overexpression of mutant p53 can promote cell invasion, we next performed a cell invasion assay and observed a significant increase in the invasive capacity of mutant p53-expressing cells compared with empty vector-transfected control cells (Figure 1c). These findings were supported by concomitantly enhanced expression of metastatic-associated genes osteopontin, MMP-2 and MMP-9 in HEC-50 cells containing the p53 mutants (Supplementary Figure S1A).
Mutant p53 GOF contributes to EMT in EC cells. (a) Morphology of endometrial cancer HEC-50 cells containing a control vector or mutant p53 R175H. Scale bars represent 100 μm. (b) Protein expression of p53 and EMT markers as analyzed by immunoblot. (c) Invasion of HEC-50 cells following overexpression of mutant p53s (mean±s.d.; n=3; *P<0.01). Representative images of invaded cells are shown. (d) Images indicate mammosphere formation in HEC-50 cells expressing the indicated constructs. The number of spheres obtained from 1000 cells at 12 days after plating (scale bar=50 μm; mean±s.d.; n=3; *P<0.01). (e) Mutant R175H- or empty vector-transfected HEC-50 cells were treated with paclitaxel (0, 25, 50 and 75 nmol/l) for 48 h. Cell viability were determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (mean±s.d.; n=3; *P<0.01). (f) Relative mRNA expression of stemness markers (normalized to GAPDH) in HEC-50 cells transfected with control or R175H vector, determined by qRT–PCR (mean±s.d.; n=4; *P<0.01).
Recently, EMT has been shown to have critical roles in modulating the cancer stem-like cell phenotype and conferring increased drug resistance of cancer cells.12 To test the roles of mutant p53 GOF in acquiring stemness and drug-resistant properties in EC cells, we investigated the effects of stable expression of mutant p53 R175H on the self-renewal potential of cells using a sphere formation assay. We also assessed the chemosensitizing properties of this cell line after treatment with paclitaxel using the Cell Counting Kit-8. We found that transfection of this mutant, but not empty vector, enabled HEC-50 cells to form floating spheres in a serum-free medium (Figure 1d) and became more resistant to paclitaxel treatment (Figure 1e). To further explore the mechanisms of mutant p53 GOF-mediated cancer stemness and drug resistance, Quantitative reverse transcription (qRT–PCR) was performed to show that the mRNA levels of well-characterized stem cell markers (CD133, KLF4 and NANOG) and chemoresistance-related genes (MDR-1 and MRP-1) was highly enhanced in R175H-expressing cells (Figure 1f).
Similar to the findings obtained from stable transfection experiments, transient transfection of a vector encoding mutant p53 R248Q, but not empty vector, promoted cell invasion (Supplementary Figure S2C). In addition, this mutant also promoted EMT-like changes, including enhanced expression of ZEB1, BMI-1, N-cadherin and Vimentin, as well as repression of E-cadherin in HEC-50 cells (Supplementary Figure S2A). Taken together, these observations suggested a crucial role of mutant p53 GOF in driving EMT and invasive phenotypes of EC cells.
Knockdown of mutant p53 in EC cells causes a reversal of EMT and inhibition of cell invasion ability
To further examine whether loss of endogenous mutant p53 can inhibit EMT features, we performed shRNA-mediated knockdown of mutant p53 in HEC-1 cells, which express endogenous mutant p53 R248Q. Silencing of mutant p53 resulted in significant changes in cell morphology, and the scattered, mesenchymal-like HEC-1 cells began to exhibit a more epithelial-like cobblestone appearance (Figure 2a). Downregulation of this p53 mutant increased the expression of epithelial marker E-cadherin and repressed the expression of mesenchymal markers Twist, ZEB1, BMI-1, Snail, N-cadherin and Vimentin (Figure 2b). In agreement with these findings, knockdown of mutant p53 markedly reduced cell invasion (Figure 2c) and reduced the expression of osteopontin, MMP-2 and MMP-9 (Supplementary Figure S1B). To investigate if reduction of mutant p53 expression can suppress cancer stem-like and drug resistance properties, a sphere formation assay and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay were used. We found that HEC-1 cells transfected with p53 shRNA displayed decreased sphere formation (Figure 2d) and were more sensitive to paclitaxel (Figure 2e). These results were supported by qRT–PCR experiments in which the mRNA expression of CD133, KLF4, NANOG, MDR-1 and MRP-1 were significantly attenuated following knockdown of endogenous p53 using shRNA (Figure 2f). Collectively, these results demonstrate that knockdown of mutant p53 can reverse the EMT phenotype and rescue cell invasion of EC cells.
Knockdown of mutant p53 in EC cells causes a reversal of EMT and inhibition of cell invasion ability. (a) Morphology of endometrial cancer HEC-1 cells transfected with control shRNA vector or p53 shRNA vector (scale bar=100 μm). (b) Protein levels of p53 and EMT markers as analyzed by western blot. (c) Invasion of HEC-1 cells after p53 shRNA transfection (mean±s.d.; n=3; *P<0.01). Representative images of invaded cells are shown. (d) Images show mammosphere formation in HEC-1 cells after p53 silencing by shRNA. Number of spheres obtained from 1000 cells at 12 days after plating (scale bar=50 μm; mean±s.d.; n=3; *P<0.01). (e) Control- or p53 shRNA-transfected HEC-1 cells were treated with paclitaxel (0, 15, and 30 nmol/l) for 48 h. Cell viability was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (mean±s.d.; n=3; *P<0.01). (f) Relative mRNA expression of stemness markers (normalized to GAPDH) in HEC-1 cells after p53 silencing, determined by qRT–PCR (mean±s.d.; n=4; *P<0.01).
ZEB1 is a key downstream mediator in p53 GOF mutant-induced EMT
Previous studies have shown that ZEB1 has a crucial role in the promotion of EMT and cancer stem cell properties in human cancer cells.13 Overexpression of ZEB1 has been detected in aggressive EC.14 Therefore, induction of ZEB1 expression by a p53 GOF mutant and reduction of its expression after mutant p53 silencing allowed us to postulate that ZEB1 may be essential for p53 GOF mutant-induced EMT in EC cells. Transfection of HEC-50 cells with ZEB1 siRNA inhibited mutant p53 R175H-induced BMI-1 and Snail expression, restored E-cadherin expression (Supplementary Figure S3A) and greatly impaired p53 R175H-mediated cell invasion (Supplementary Figure S3B). However, this treatment did not significantly affect the protein level of Twist. Following transfection with ZEB1 siRNA in HEC-1 cells, the mRNA expression of BMI-1 and Snail was suppressed and E-cadherin was elevated (Supplementary Figure S3C). These data indicate that ZEB1 acts as an important downstream effector of these p53 mutants to mediate the EMT process in EC cells.
The p53 GOF mutants contribute to global repression of miRNA expression
To identify miRNAs mediated by the p53 GOF mutants, we performed array-based miRNA profiling of HEC-50 cells transduced with either p53 mutants or empty vector. Of 188 human miRNAs assayed, 23 miRNAs were expressed above background levels. Ectopic overexpression of mutant p53 R273H, R175H and C135Y in HEC-50 cells led to a global downregulation of all these miRNAs (Supplementary Figure S4A). We further validated the microarray results using qRT–PCR (Figure 3b). Notably, the expression of several miRNAs with known tumor suppressor activity, including let-7b,3 miR-143,15 miR-194,16 miR-424,17 miR-451,18 and miR-14619 were significantly reduced in p53-mutant-expressing cells (Supplementary Figure S4B). Thus, these results suggest that global repression of miRNA expression is likely to be a critical mechanism for p53 GOF mutant-enhanced EC tumorigenesis.
Mutant p53 binds to and transrepresses the promoter of miR-130b. (a) Schematic of algorithm used to select candidate microRNAs that potentially target ZEB1, and are negatively regulated by mutant p53s. (b, c) Relative miR-130b expression levels in HEC-50 cells transfected with mutant p53 vector (b), or in HEC-1 cells after p53 silencing by shRNA (c), were determined by qRT–PCR (mean±s.d.; n=4; *P<0.01). (d) Location and sequence of predicted p53-binding sites in the promoter of miR-130b gene. Mutated residues (red) are indicated at the bottom. (e) ChIP–qPCR analysis of mutant p53 (DO-7 antibody) binding to the miR-130b promoter region in HEC-50 cells. Human telomerase (hTERT) was used as a positive control. The fold enrichment over the IgG control is represented (mean±s.d.; n=3; *P<0.01). (f) HEC-50 cells were transfected with luciferase reporter plasmid pGL3-130b or empty pGL3-basic vector, along with control vector, wild-type p53 or mutant p53 R175H vector, and relative luciferase activity were assayed (mean±s.d.; n=3; *P<0.01). All qRT–PCR or luciferase values were normalized to GAPDH or Renilla activity, respectively.
Mutant p53 binds to and transrepresses the promoter of miR-130b
Considering the important roles of ZEB1 in regulating EMT, we next sought to determine whether any of the 23 miRNAs may target ZEB1, and if repression of these miRNAs by mutant p53 may contribute to increased ZEB1 expression in EC cells. We first searched for all predicted miRNA-ZEB1 interactions by using two target-prediction algorithms: TargetScan (http://www.targetscan.org) and microRNA.org (http://www.microrna.org), and detected four miRNAs that potentially bind to the 3′ untranslated regions (3′-UTR) of ZEB1 mRNA. We then cross-referenced these four miRNAs with the 23 miRNAs identified by miRNA microarray. We found three miRNAs (miR-130b, miR-301a and miR-301b) that were downregulated in p53-mutant-expressing cells and predicted to bind to ZEB1 3′-UTR (Figure 3a). We selected miR-130b to investigate its effects on EMT and EC cell invasion because transfection of miR-301a and miR-301b did not substantially alter the protein expression of ZEB1 in EC cells (data not shown).
To evaluate whether the p53 mutants (R273H, R175H and C135Y) control the expression of miR-130b, we examined the effects of overexpression of mutant p53 on the expression of miR-130b in HEC-50 cells. The qRT–PCR analysis confirmed a significant decrease in mature miR-130b levels following transfection with the p53 mutants (Figure 3b). Similarly, the p53 mutation R248Q, but not the empty vector, inhibited the expression of miR-130b when expressed transiently (Supplementary Figure S2B). On the other hand, HEC-1 cells transfected with p53 shRNA exhibited a marked elevation in the level of miR-130b (Figure 3c). These results suggest that the endogenous expression of miR-130b is negatively regulated by p53 mutants.
Recent evidence has established an association between p53 and several miRNAs, such as miR-34,20 miR-19221 and miR-200c.5 Transcription of these miRNAs is directly regulated by p53. In particular, several studies have suggested that miR-200c is downregulated in EC tissues,22 and restoration of miR-200c expression in HEC-50 cells decreases cell invasion.23 Using qRT–PCRs to compare miRNA levels in HEC-50 cells, we found that overexpression of mutant p53 R175H and C135Y decrease the expression of miR-200c by 30–20%, whereas mutant R273H has no effects on its expression (Supplementary Figure S5), indicating that downregulation of miR-200c is involved in mutant p53 GOF-induced EC cell invasion.
However, it remains unknown whether p53 mutants function as a transcription regulator of miR-130b. Therefore, we searched for p53-binding sites in the miR-130b promoter using a bioinformatics approach.24 Importantly, we found a conserved p53-binding site (5′-GGGCATGGTGGCTCATGCCT-3′) with a ranking score of 83 (Figure 3d). To determine whether an endogenous p53 mutant can bind this site, chromatin immunoprecipitation (ChIP)–qPCR analysis was performed on HEC-1 cells. The human telomerase (hTERT) promoter served as a positive control, as it has been previously shown that p53 mutants can bind this promoter.25 Both miR-130b (sixfold) and hTERT (eightfold) promoter sequences were specifically enriched by anti-p53 antibodies, but not by non-specific antibodies (Figure 3e). These data suggest that miR-130b is a direct target of mutant p53 in EC cells.
To assess if the downregulation of miR-130b expression is mediated by transrepression of the p53 mutants, we cloned the p53-binding sequence of the miR-130b promoter upstream of firefly luciferase to yield a WT plasmid pGL3-130b, and further generated mutant pGL3-130b luciferase vectors containing mutations in the candidate p53-binding site. The WT pGL3-130b or mutant pGL3-130b vector was transfected into HEC-50 cells with either a control vector, mutant p53 R175H or WT p53. Interestingly, the luciferase activity of WT pGL3-130b was significantly repressed by R175H, but was transactivated by WT p53. However, expression of mutant p53 or WT p53 did not affect the luciferase activity of mutant pGL3-130b (Figure 3f). Therefore, our observations by qRT–PCR, ChIP–qPCR and the luciferase assay collectively demonstrate that a GOF p53 mutant binds to and transrepresses the miR-130b promoter.
Our results showing a fivefold increase in the ability of WT p53 to transactivate the promoter of miR-130b (Figure 3f, lane 2) raised an interesting possibility that WT p53 controls metastasis through modulation of miR-130b. Therefore, we transiently transfected the WT p53 expression vector into HEC-50 cells (Figure 4a). A qRT–PCR analysis revealed that expression of WT p53 protein significantly induced the levels of miR-130b and also slightly increased the expression of miR-200c (Figure 4b), which is a known target of WT p53.6 In WT p53-expressing HHUA cells, activation of p53 in response to the Mdm2 antagonist Nutlin-3 (Figure 4c) enhanced the level of miR-130b, but this was abolished by the shRNA-mediated knockdown of p53 (Figure 4d). Consistent with an earlier report,6 the knockdown of WT p53 in HHUA cells induced changes associated with EMT, such as a mesenchymal morphology (Figure 4e), low expression of E-cadherin, upregulation of ZEB1 and BMI-1 (Figure 4f), and increased cell invasion (Figure 4g). We also observed an enrichment of WT p53 binding to both the miR-130b and p21 promoters using a ChIP–PCR analysis (Figure 4h). In addition, WT p53 was able to transactivate a pGL-3-miR-130b luciferase reporter gene (Figure 4i). Silencing of p53 by shRNA abrogated Nutlin-3-stimulated luciferase activities of the miR-130b promoter (Figure 4i, compare lane 5 to lanes 7 and 8). These data indicated that mutant and WT p53 exert opposite effects on miR-130b expression, which supports the hypothesis that a p53 GOF mutant contributes to EC carcinogenesis by altering the expression of miR-130b.
WT p53 transactivates the promoter of miR-130b. (a) WT p53 protein level in HEC-50 cells transfected with WT p53 expression vector or control vector. (b) qRT–PCR for miR-130b and miR-200c in HEC-50 cells transfected with WT p53 expression vector or control vector (mean±s.d.; n=4; *P<0.01). (c, d) HHUA cells transfected with p53 shRNA vector or control vector were treated with 5 μmol/l of Nutlin-3 or dimethyl sulfoxide (DMSO) for 12 h. WT p53 protein (c) and miR-130b expression (d) were detected by western blot analysis and qRT–PCR (mean±s.d.; n=4; *P<0.01), respectively. (e) Morphology of HHUA cells after p53 silencing. Scale bars represent 200 μm. (f) Western blot analysis for EMT markers in HHUA cells after p53 silencing. (g) Invasion assay of HHUA cells after transfection with p53 shRNA (mean±s.d.; n=3; *P<0.01). (h) ChIP–qPCR analysis of WT p53 (DO-7 antibody) binding to the miR-130b promoter region in HHUA cells. p21 was used as a positive control. The fold enrichment over the IgG control is represented (mean±s.d.; n=3; *P<0.01). (i) Indicated HHUA cells were transfected with luciferase reporter plasmid pGL3-130b or empty pGL3-basic vector, and treated with 5 μmol/l of Nutlin-3 or DMSO for 12 h. Relative luciferase activity was determined (mean±s.d.; n=3; *P<0.01). All qPCR or luciferase values were normalized to GAPDH or Renilla activity, respectively.
Despite direct transcriptional regulation by mutant p53, some GOF effects of the p53 mutants may depend on their ability to inactivate p53 family members p63 or p73.26 Furthermore, p63 has been shown to inhibit metastasis through transactivation of miR-130b.27 Therefore, we determined whether p63 inhibition by the p53 mutants is involved in the p53 GOF mutant-induced suppression of miR-130b in HEC-50 cells. We found that downregulation of p63 protein expression by p63 siRNA (Supplementary Figure S6A) resulted in a dose-dependent decrease in p21 luciferase activity (Supplementary Figure S6C). As expected, transient transfection of the WT p53 expression vector markedly transactivated the p21 promoter (Supplementary Figure S6C). However, the mRNA expression of miR-130b did not substantially change after p63 knockdown (Supplementary Figure S6B). Thus, p63 inhibition is not likely to be responsible for p53 GOF mutant-induced suppression of miR-130b in EC cells.
miR-130b impairs cell invasion by targeting ZEB1
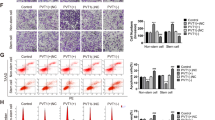
To investigate whether miR-130b can control EC cell invasion through the modulation of ZEB1 expression, we used TargetScan and miRviewer to search for miRNA-binding sites in the 3'-UTR of ZEB1. The miR-130b was found to possess five evolutionary-conserved binding sites, suggesting that a potential interaction between miR-130b seed region and ZEB1 mRNA 3′-UTR (Figure 5a). To test if miR-130b binds directly to ZEB1 transcript, we transfected biotin-labeled miR-130b into HEC-50 cells, pulled down mRNAs bound to miR-130b and quantified ZEB1 transcript using qRT–PCRs. We found that the levels of ZEB1 mRNA were highly enriched by miR-130b pull-down, as compared with control transcripts of housekeeping genes 5S rRNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Supplementary Figure S7). Real-time PCR and western blot analysis demonstrated that, in HEC-50 cells with high levels of ZEB1, restoration of miR-130b reduced ZEB1 mRNA and protein expression (Figures 5b and c). In contrast, in HEC-1 cells, which express low levels of ZEB1, miR-130b inhibition by transfection with anti-miR-130b increased ZEB1 mRNA and protein levels (Figures 5b and c). These results demonstrate that miR-130b directly interacts with ZEB1 mRNA and represses its expression.
miR-130b impairs cell invasion by targeting ZEB1. (a) Schematic representation of the 3′-UTR of ZEB1 with the predicted target site for miR-130b. Sequence of mature miR-130b reveals the evolutionary conservation of the target site across five species (below). (b, c, e) qRT–PCR (b, mean±s.d.; n=3; *P<0.05), western blotting (c) and cell invasion assay (e, mean±s.d.; n=3; *P<0.01) of HEC-50 or HEC-1 cells transfected with pre-miR-130b or anti-miR-130b, respectively. (d) Reporter constructs containing either wild-type ZEB1 3′-UTR or ZEB1 3′-UTR with mutation at the predicted miR-130b target sequence were co-transfected into HEC-50 cells, along with miR-130b, control miRNA, anti-miR-130b or control anti-miRNA. Relative luciferase activity was assayed (mean±s.d.; n=3; *P<0.01). (f) Expression of EMT and stemness markers in HEC-50 cells transfected with pre-130b or control miRNA were analyzed by qRT–PCR (mean±s.d.; n=4; *P<0.01). All qPCR or luciferase values were normalized to GAPDH or Renilla activity, respectively.
To assess if miR-130b targets ZEB1 3′-UTR, reporter constructs containing either the WT ZEB1 3′-UTR or ZEB1 3′-UTR with mutation at the predicted miR-130b target sequence were co-transfected into HEC-50 cells together with miR-130b, control miRNA, anti-miR-130b or control anti-miRNA. Transduction of miR-130b caused marked inhibition of the WT ZEB1 3′-UTR, but had no effect on mutant ZEB1 3′-UTR (Figure 5d, compare lane 1 to lanes 2 and 3). In addition, miR-130b inhibition by anti-miR-130b substantially increased luciferase activities of WT ZEB1 3′-UTR compared with control anti-miRNA (Figure 5d, compare lane 4 to lane 5). These data together suggest that miR-130b downregulates ZEB1 expression in EC cells by destabilizing the ZEB1 mRNA as well as translational suppression.
To determine the functional effects of miR-130b-mediated ZEB1 suppression on cell invasion, a cell invasion assay was performed. We found that elevated expression of miR-130b in HEC-50 cells decreased cell invasion, and knockdown of miR-130b by anti-miR-130b in HEC-1 cells enhanced cell invasion (Figure 5e). In agreement with these findings, transfection of miR-130b, but not control miRNA, significantly reduced the mRNA levels of BMI-1, Snail, KLF4, NANOG and MDR-1, and increased mRNA expression of E-cadherin in HEC-50 cells (Figure 5f). Taken together, these results suggest that miR-130b directly targets ZEB1, and as a result reverses EMT-associated EC cell invasion.
The p53 GOF mutants stimulate EMT features through downregulation of miR-130b
To further define the involvement of miR-130b in mutant p53-stimulated ZEB1 expression and EMT characteristics, miR-130b was transfected into mutant p53 R175H-expressing HEC-50 cells. Reintroduction of miR-130b abolished the mRNA expression of ZEB1, Snail, BMI-1, KLF4 and NANOG, restored E-cadherin expression and markedly diminished p53R175H-induced cell invasion (Figures 6a and b). To further confirm these results, we used HEC-1 cells expressing shRNA against p53 or control cells to show that transfection with anti-miR-130b was capable of restoring the mRNA levels of ZEB1, Snail, BMI-1, KLF4 and NANOG, as well as decrease the expression of E-cadherin and initiate sphere formation (Figures 6c and d). These data demonstrate that a p53 GOF mutant downregulates miR-130b expression, which results in activation of ZEB1, and its downstream pathway and contributes to the induction of EMT and increased EC cell invasion.
The p53 GOF mutants stimulate EMT features through downregulation of miR-130b. (a, c) qRT–PCR for EMT and stemness markers in HEC-50 cells (a) or in HEC-1 cells (c) expressing indicated constructs, and pre-miRNAs and anti-miRNAs (mean±s.d.; n=3; *P<0.01). (b) Invasion assay of HEC-50 cells expressing indicated vectors and pre-miRNAs (mean±s.d.; n=3; *P<0.01). (d) Sphere formation assay of HEC-1 cells expressing indicated vectors and anti-miRNAs (mean±s.d.; n=3; *P<0.01).
Clinical association of miR-130b expression with prognosis of EC patients
The expression of miR-130 was significantly reduced (P=0.02) in EC tissues (Figure 7a). Moreover, patients with higher expression levels of miR-130b survived longer (P=0.05) than patients with lower expression levels (Figure 7b).
Discussion
In this study, we have demonstrated that a GOF p53 mutant can induce EMT and increase invasive properties in EC cells by regulating a large set of EMT-associated genes. More importantly, we provide an underlying mechanism for p53 GOF-enhanced metastasis: mutant p53 can bind directly and transrepress the promoter of miR-130b, which is a specific inhibitor of ZEB1, leading to the upregulation of ZEB1 and subsequent activation of the E-cadherin suppressors BMI-1 and Snail (Figure 8). Notably, the effects of mutant p53 on EMT features and cell invasion can be partly abolished by restoration of miR-130b expression. Therefore, re-expression of miR-130b may inhibit tumor metastasis and progression, providing a potential therapeutic use in patients with EC.
Although upregulation of some miRNAs is reported in different tumors,28 the widespread reduction of miRNA expression has been observed in a range of tumor types and is associated with increased metastatic activity.29, 30 Our findings suggest that the principal consequence of a p53 GOF mutant on miRNA expression is global repression. Thus, the correlation between a p53 GOF mutant and general repression of miRNAs suggests that the overall repression of miRNAs, especially those miRNAs with tumor suppressor function, is involved in p53 GOF mutant-stimulated EC metastasis and progression.
Several reports showed that p53 mutations not only target a set of genes that are different from those controlled by WT p53 such as c-myc,31 but also differently modulate WT p53-target genes.32, 33, 34 Mechanistically, the transcriptional effects of mutant p53 on WT p53-dependent promoters are mediated by at least four types of molecular interactions: (1) mutant p53 binds to WT p53-target gene promoters in the region distinct from WT p53 response elements.35 (2) However, ChIP analysis reveals that mutant p53 physically associates with several promoters, such as EGR136 and MSP/MST-1,37 which also interact with WT p53. (3) There is increasing evidence that both WT and mutant p53 can form a complex with sequence-specific transcription factors (Sp1,38, 39 NF-Y,40, 41 or other factors), and be recruited to binding sites of those factors on chromatin, and modulate their transcriptional activities. (4) In addition, p53 mutants can display the WT p53 conformation due to a shift in the conformational equilibrium,42 and thereby bind to the consensus sequence.43 Our data indicate that both mutant p53 and WT p53 are present on the miR-130b promoter to regulate its expression, which is consistent with the reported finding of mutant p53. Future studies will be required to identify the transcriptional factors that specifically interact with mutant p53 and clarify the biological significance of their physical interaction.
Previous studies have shown that miR-130b is downregulated in hepatocellular carcinoma44 and aggressive papillary thyroid carcinoma.45 Furthermore, downregulation of miR-130b confers a multidrug-resistant phenotype in ovarian cancer cells.46 However, other reports also suggest that overexpression of miR-130b in CD133 (+) liver tumor-initiating cells increased their self-renewal capacity and chemoresistance.47 We found that the transcriptional inactivation of miR-130b by mutant p53 is required for p53 GOF mutant-mediated EMT and invasive phenotypes in EC cells. These results suggest that miR-130b may have a dual function as both a tumor suppressor and oncogene, depending on the cancer type and cellular context.
The HEC-50 cell line (p53 null) was derived from a patient with invasive grade 3 EC. Thus, the inverse relationship between miR-130b and mutant p53 expression observed in these cells, which is a genetic alteration frequently found in aggressive EC, suggests that miR-130b expression is likely reduced in later stages of tumor progression when mutant p53 becomes the main driver of invasion and metastasis.
Some evidence has suggested that miR-194 is a WT p53-responsive miRNA with potent anti-proliferative activity.48 Interestingly, we have recently shown that miR-194 is able to inhibit EMT and cell invasion of EC cells by targeting oncogene BMI-1.16 Our present study suggests that the expression of miR-194 is negatively regulated by mutant p53 in EC cells. The significance and clinical relevance of miR-130b were further demonstrated in EC patients (Figure 7). Therefore, we postulate that p53 mutations induce EMT and promote EC metastasis, at least in part, through regulating both miR-130b/ZEB1 and miR-194/BMI-1 signaling pathways (Figure 8). Our results open a possibility that multiple molecular mechanisms with different miRNAs are involved in p53 GOF mutant-dependent EMT programming. Further research is clearly needed to understand the mechanisms of p53 mutant-mediated EMT induction and the functional cross-talk between p53 signal pathways and miRNA-modulated gene expression profiles.
Materials and methods
Cell culture
The EC cell lines HEC-50 and HEC-1 were cultured in Eagle’s MEM medium (Sigma-Aldrich, Poole, UK) supplemented with 15% fetal bovine serum. The EC cell line HHUA was maintained in Ham’s F12 medium containing 15% fetal bovine serum. All cell lines used were obtained from the RIKEN cell bank (Tsukuba, Japan).
Generation of cells overexpressing mutant p53 and knockdown of WT p53
HEC-50 cells at 80% confluency were transfected with vectors containing WT p53, mutant p53 (R273H, R175H, C135Y) or control vector as previously described.49 The selection of stably transfected clones was achieved using a medium containing 400 μg/ml of G418 (Sigma-Aldrich) in the media. We knocked down p53 expression in HEC-1 and HHUA cells using a pSUPER-p53 vector or pSUPER control vector performed as previously described,49 and selected cells with 1 μg/ml puromycin (Sigma-Aldrich).
Western blot analysis
Whole-cell lysates were obtained using the M-Per Mammalian Protein Extraction Reagent (Pierce Biotechnology, Rockford, IL, USA). Proteins (40 μg) were separated on 10% SDS–PAGE and transferred to nitrocellulose membranes. Antigen–antibody complexes were detected using the electrochemiluminesence blotting analysis system (Amersham Pharmacia Biotech, London, UK). The following antibodies were used: mouse monoclonal anti-p53 (DO-7), goat polyclonal anti-ZEB1 (C-20), rabbit polyclonal anti-Twist (sc-81417), goat polyclonal anti-Snail (sc-10432), mouse monoclonal anti-p63 (sc-8431) and mouse monoclonal anti-GAPDH (sc-47724) (Santa Cruz, Santa Cruz, CA, USA). Rabbit polyclonal anti-BMI-1 (ab38295) (Abcam, Cambridge, MA, USA), rabbit polyclonal anti-E-cadherin (A01589), rabbit polyclonal anti-Vimentin antibody (A01189) (GenScript, Edison, NJ, USA) and mouse monoclonal anti-N-cadherin (BD Transduction, San Jose, CA, USA) antibodies were also used. Primary and secondary antibodies were used at 1:1000 and 1:5000 dilutions, respectively.
Molecular cloning of miR-130b promoter
The genomic region overlapping the p53-binding site was synthesized with forward primer (5′-ATACGCGTGGGTAAGGGACTCCTGAAGC-3′, Mlu I) and reverse primer (5′-CGAGATCTGAGACAAGGTTTCACCACGTT-3′, BglII), and sub-cloned into MluI/BglII sites of the pGL3-basic plasmid (Promega, Madison, WI, USA) to produce pGL3-miR-130b (WT pGL3-miR-130b). (The underlined primer sequence indicates the sites for enzyme MluI.) Correct insertion was confirmed by gel electrophoresis and DNA sequencing.
ChIP assay and qPCR analysis
The ChIP assay was performed using the Pierce Agarose ChIP kit (Pierce; Thermo Scientific, Rockford, IL, USA) according to the manufacturer’s protocol. Immunoprecipitation was carried out using mouse monoclonal p53 antibody (DO-7) or unrelated rabbit IgG as a negative control. To amplify the potential p53-binding site from nucleotides −3540 to −3520 in the promoter of miR-130b, real-time PCR was performed using the forward primer 5′-TTCATCCGTTCTCACACTGC-3′ and the reverse primer 5′-CAGGCTGGTCTCGAACTCC-3′. The human telomerase (hTERT) and p21 genes were used as positive controls for mutant p53 and WT p53 binding, respectively.21, 25
Patients and samples
The clinical sample cohort used for this study was approved by the Institutional Review Board of Stony Brook Medicine. Written informed consent was obtained from all participants involved in the study. Paraffin blocks containing formalin-fixed paraffin-embedded tissue samples were acquired from the archived collections of the Department of Pathology and used for subsequent analyses. The specimens were selected from samples obtained between 1995 and 2010, and each case had up to 15 years of clinical follow-up information. For RNA extraction, tumor samples and the adjacent normal tissues were obtained from 32 EC patients who underwent hysterectomy at Stony Brook Medicine, Stony Brook, New York. The characteristics of these patients are shown in Table 1.
RNA isolation
Using archived containing formalin-fixed paraffin-embedded tissues, separate areas of tumor and normal endometrium were identified from the corresponding hematoxylin and eosin-stained sections, and cores measuring 1.5 mm in diameter and 2 mm in length (∼0.005 g) were extracted. The samples were then deparaffinized, hydrated and digested with proteinase K. Subsequently, total RNA was isolated using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA).
Real-time qRT–PCR analysis of miRNA expression
The miR-130b-specific primers and the internal control RNU44 gene were purchased from Ambion (Applied Biosystems, Foster City, CA, USA). cDNA synthesis was performed using the High Capacity cDNA Synthesis Kit (Applied Biosystems). qRT–PCR was carried out on an Applied Biosystems 7500 Real time system (ABI 7500HT instrument) using the TaqMan Gene Expression Assay.
Statistical analysis
All experiments were performed in triplicate. All statistical analyses were performed using GraphPad Prism software 5.0 (GraphPad Software, Inc., San Diego, CA, USA) and SPSS statistical software (SPSS Japan Inc., Tokyo, Japan). A Student’s t-test was used for analysis, and statistical significance was defined as P<0.05. Gene expression ΔCt values of miR-130b from each sample were calculated by normalizing them to the expression of the RNU44 internal control, and relative quantification values were plotted. The differences between tumor and normal tissues were analyzed using the Wilcoxon matched pairs test. Kaplan–Meier survival curves were generated to evaluate the correlation of miR-130b expression levels with survival rate.
References
Kang Y, Massagué J . Epithelial-mesenchymal transitions: twist in development and metastasis. Cell 2004; 118: 277–279.
Korpal M, Lee ES, Hu G, Kang Y . The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 2008; 283: 14910–14914.
Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res 2009; 69: 6704–6712.
Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove R et al. miR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology 2010; 52: 2148–2157.
Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med 2011; 208: 875–883.
Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW et al. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 2011; 13: 317–323.
Rivlin N, Brosh R, Oren M, Rotter V . Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer 2011; 2: 466–474.
Scian MJ, Stagliano KE, Anderson MA, Hassan S, Bowman M, Miles MF et al. Tumor-derived p53 mutants induce NF-kappaB2 gene expression. Mol Cell Biol 2005; 25: 10097–10110.
Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J et al. Prognostic values of microRNAs in colorectal cancer. Biomark Insights 2006; 2: 113–121.
Kogan-Sakin I, Tabach Y, Buganim Y, Molchadsky A, Solomon H, Madar S et al. Mutant p53 (R175H) upregulates Twist1 expression and promotes epithelial-mesenchymal transition in immortalized prostate cells. Cell Death Differ 2011; 18: 271–281.
Fujisawa T, Watanabe J, Kamata Y, Hamano M, Hata H, Kuramoto H . VEGF expression and its reguration by p53 gene transfection in endometrial carcinoma cells. Hum Cell 2003; 16: 47–54.
Singh A, Settleman J . EMT cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 2010; 29: 4741–4751.
Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009; 11: 1487–1495.
Singh M, Spoelstra NS, Jean A, Howe E, Torkko KC, Clark HR et al. ZEB1 expression in type I vs type II endometrial cancers: a marker of aggressive disease. Mod Pathol 2008; 21: 912–923.
Clapé C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, Iborra F et al. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS One 2009; 4: e7542.
Dong P, Kaneuchi M, Watari H, Hamada J, Sudo S, Ju J et al. MicroRNA-194 inhibits epithelial to mesenchymal transition of endometrial cancer cells by targeting oncogene BMI-1. Mol Cancer 2011; 10: 99.
Forrest AR, Kanamori-Katayama M, Tomaru Y, Lassmann T, Ninomiya N, Takahashi Y et al. Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation. Leukemia 2010; 24: 460–466.
Bandres E, Bitarte N, Arias F, Agorreta J, Fortes P, Agirre X et al. MicroRNA-451 regulates macrophage migration inhibitory factor production and proliferation of gastrointestinal cancer cells. Clin Cancer Res 2009; 15: 2281–2290.
Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR . Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res 2009; 69: 1279–1283.
Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 2007; 26: 731–743.
Song B, Wang Y, Kudo K, Gavin EJ, Xi Y, Ju J . miR-192 Regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin Cancer Res 2008; 14: 8080–8086.
Castilla MÁ, Moreno-Bueno G, Romero-Pérez L, Van De Vijver K, Biscuola M, López-García MÁ et al. Micro-RNA signature of the epithelial-mesenchymal transition in endometrial carcinosarcoma. J Pathol 2011; 223: 72–80.
Cochrane DR, Spoelstra NS, Howe EN, Nordeen SK, Richer JK . MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol Cancer Ther 2009; 8: 1055–1066.
Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J . Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res 2006; 12: 2014–2024.
Scian MJ, Stagliano KE, Deb D, Ellis MA, Carchman EH, Das A et al. Tumor-derived p53 mutants induce oncogenesis by transactivating growth-promoting genes. Oncogene 2004; 23: 4430–4443.
Oren M, Rotter V . Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2010; 2: a001107.
Su X, Chakravarti D, Cho MS, Liu L, Gi YJ, Lin YL et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 2010; 467: 986–990.
Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z . Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One 2009; 4: e8003.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D et al. MicroRNA expression profiles classify human cancers. Nature 2005; 435: 834–838.
Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA 2008; 105: 7004–7009.
Frazier MW, He X, Wang J, Gu Z, Cleveland JL, Zambetti GP . Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol Cell Biol 1998; 18: 3735–3743.
Sun Y, Cheung JM, Martel-Pelletier J, Pelletier JP, Wenger L, Altman RD et al. Wild type and mutant p53 differentially regulate the gene expression of human collagenase-3 (hMMP-13). J Biol Chem 2000; 275: 11327–11332.
Chin KV, Ueda K, Pastan I, Gottesman MM . Modulation of activity of the promoter of the human MDR1 gene by Ras and p53. Science 1992; 255: 459–462.
Kim E, Deppert W . Transcriptional activities of mutant p53: when mutations are more than a loss. J Cell Biochem 2004; 93: 878–886.
Zalcenstein A, Stambolsky P, Weisz L, Müller M, Wallach D, Goncharov TM et al. Mutant p53 gain of function: repression of CD95(Fas/APO-1) gene expression by tumor-associated p53 mutants. Oncogene 2003; 22: 5667–5676.
Weisz L, Zalcenstein A, Stambolsky P, Cohen Y, Goldfinger N, Oren M et al. Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res 2004; 64: 8318–8327.
Zalcenstein A, Weisz L, Stambolsky P, Bar J, Rotter V, Oren M . Repression of the MSP/MST-1 gene contributes to the antiapoptotic gain of function of mutant p53. Oncogene 2006; 25: 359–369.
Gualberto A, Baldwin AS . p53 and Sp1 interact and cooperate in the tumor necrosis factor-induced transcriptional activation of the HIV-1 long terminal repeat. J Biol Chem 1995; 270: 19680–19683.
Chicas A, Molina P, Bargonetti J . Mutant p53 forms a complex with Sp1 on HIV-LTR DNA. Biochem Biophys Res Commun 2000; 279: 383–390.
Imbriano C, Gurtner A, Cocchiarella F, Di Agostino S, Basile V, Gostissa M et al. Direct p53 transcriptional repression: in vivo analysis of CCAAT-containing G2/M promoters. Mol Cell Biol 2005; 25: 3737–5371.
Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A et al. Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell 2006; 10: 191–202.
McLure KG, Lee PW . p53 DNA binding can be modulated by factors that alter the conformational equilibrium. EMBO J 1999; 18: 763–770.
Zhang W, Funk WD, Wright WE, Shay JW, Deisseroth AB . Novel DNA binding of p53 mutants and their role in transcriptional activation. Oncogene 1993; 8: 2555–2559.
Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem 2006; 99: 671–678.
Yip L, Kelly L, Shuai Y, Armstrong MJ, Nikiforov YE, Carty SE et al. MicroRNA signature distinguishes the degree of aggressiveness of papillary thyroid carcinoma. Ann Surg Oncol 2011; 18: 2035–2041.
Yang C, Cai J, Wang Q, Tang H, Cao J, Wu L et al. Epigenetic silencing of miR-130b in ovarian cancer promotes the development of multidrug resistance by targeting colony-stimulating factor 1. Gynecol Oncol 2012; 124: 325–334.
Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A et al. miR-130b promotes CD133 (+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell 2010; 7: 694–707.
Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T et al. p53-responsive microRNAs 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 2008; 68: 10094–10104.
Dong P, Xu Z, Jia N, Li D, Feng Y . Elevated expression of p53 gain-of-function mutation R175H in endometrial cancer cells can increase the invasive phenotypes by activation of the EGFR/PI3K/AKT pathway. Mol Cancer 2009; 8: 103–110.
Acknowledgements
We appreciate the critical review by Ms Sonya R Lorrain. This work was funded by a grant from Women’s Health Educational System, a Grant-in-Aid from the Ministry of Health, Labour and Welfare of Japan, Shanghai Municipal Natural Science Foundation (11ZR1430500) and by Stony Brook University Translational Research Laboratory Start-up fund (J Ju), R01CA155019 (J Ju) and R33CA147966 (J Ju). We thank Dr Zhujie Xu for his technical assistance.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Dong, P., Karaayvaz, M., Jia, N. et al. Mutant p53 gain-of-function induces epithelial–mesenchymal transition through modulation of the miR-130b–ZEB1 axis. Oncogene 32, 3286–3295 (2013). https://doi.org/10.1038/onc.2012.334
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.334
Keywords
This article is cited by
-
Epithelial-to-mesenchymal transition status correlated with ultrastructural features, and TP53 mutation in patient-derived oral cancer cell lines
Molecular Biology Reports (2023)
-
Transition of amyloid/mutant p53 from tumor suppressor to an oncogene and therapeutic approaches to ameliorate metastasis and cancer stemness
Cancer Cell International (2022)
-
STAT3/miR-130b-3p/MBNL1 feedback loop regulated by mTORC1 signaling promotes angiogenesis and tumor growth
Journal of Experimental & Clinical Cancer Research (2022)
-
Should mutant TP53 be targeted for cancer therapy?
Cell Death & Differentiation (2022)
-
Targeting mutant p53 for cancer therapy: direct and indirect strategies
Journal of Hematology & Oncology (2021)