Abstract
Expression of hypoxia-inducible factors (HIFs) and N-myc downstream-regulated gene 3 (NDRG3) are oxygen-dependently regulated by prolyl hydroxylase domain (PHD) enzymes. Little is known about the role of NDRG3 in the cellular adaptation to hypoxia, whereas the roles of HIFs are well understood. In this study, we investigated how NDRG3 affects the hypoxic response in prostate cancer cells. Compared with HIF-1α, hypoxic induction of NDRG3 was observed at a later phase. NDRG3 reduced hypoxic expression of HIF-1α by inhibiting AKT-driven translation of HIF1A mRNA. In addition, NDRG3 functionally inhibited HIF-1 by dissociating the coactivator p300 from HIF-1α. Accordingly, NDRG3 may fine-tune the HIF-1 signaling pathway to cope with long-term hypoxia. Of the diverse effects of HIF-1α on cancer progression, hypoxia-induced cell migration was investigated. In transwell chambers, NDRG3 negatively regulated the migration and invasion of prostate cancer cells under hypoxia. An informatics analysis using Gene Expression Omnibus (GEO) revealed that NDRG3 downregulation is associated with prostate cancer metastasis and high expression of HIF-1 downstream genes. In cancer tissue arrays, NDRG3 expression was lower in prostate cancer tissues with a Gleason score of 8 or greater and was inversely correlated with HIF-1α expression. Therefore, NDRG3 may have an anti-metastatic function in prostate cancer under a hypoxic microenvironment.
Similar content being viewed by others
Introduction
Metazoan cells maintain oxygen homeostasis through a balance between mitochondrial oxygen consumption and external oxygen supply. Disruption of this balance results in energy depletion or oxidative injury, which may lead to various diseases including cancer1. Hypoxia-inducible factor 1 and 2 (HIF-1/2), which are bHLH-PAS family transcription factors composed of α and β subunits, play key roles in maintaining oxygen homeostasis2,3.
HIF-1α expression is tightly regulated by the prolyl hydroxylases PHD 1–3 whose activities are dependent on the ambient oxygen tension. In aerobic conditions, PHDs hydroxylate the Pro-402 and Pro-564 residues on the ODD domain in HIF-1α, allowing the von Hippel-Lindau protein (pVHL) E3 ligase complex to ubiquitinate HIF-1α, promoting proteasomal degradation4,5,6. In oxygen-deficient conditions, however, HIF-1α is stabilized because PHDs are inactivated. HIF-1α dimerizes with HIF-1β/ARNT in the nucleus, leading to the expression of hundreds of downstream genes7,8. The activity of HIF-1α is also oxygen-dependently regulated by FIH-1 (factor inhibiting HIF-1), which prevents HIF-1α from binding with its co-activators CBP/p300 by hydroxylating the Asn-803 residue in the HIF-1α C-terminal transactivation domain (CAD)9,10. In addition to the oxygen-dependent regulation, HIF-1α expression is also determined at the translational step, which is activated by the PI3K–AKT–mTOR pathway. This pathway is highly activated in prostate cancer cells because of the deletion of the PTEN gene, so HIF-1α is frequently overexpressed in prostate cancer11,12.
The N-myc downstream-regulated gene (NDRG) family, which is composed of four members (NDRG1–4), is involved in hypoxia-induced reprogramming of cancer metabolism13. NDRG members display tumor-suppressive behaviors in various cancers, so their expression is suggested to be a good prognostic marker13. Recently, NDRG3 was revealed as another target for the PHD oxygen sensors14. Similar to HIF-1α, NDRG3 is prolyl-hydroxylated under normoxia by PHD2, poly-ubiquitinated by pVHL, and degraded through the proteasomal pathway. NDRG3 becomes stable under hypoxia because this degradation process is blocked. If hypoxia persists, accumulated lactate interferes with the interaction between NDRG3 and PHD2. Therefore, a lack of oxygen and lactate production both facilitate the stabilization of NDRG3 in long-term hypoxia. Functionally, NDRG3 can prolong hypoxic responses in persistent hypoxia, whereas short-lived HIF-1α participates in immediate hypoxic responses. However, we aimed to investigate whether HIF-1α and NDRG3 work cooperatively towards cellular adaptation to hypoxia. In this work, we investigated the cross-talk between HIF-1α and NDRG3 in prostate cancer cells. Furthermore, we examined the consequence of the hypoxic induction of NDRG3 in cancer metastasis.
Materials and methods
Cell culture
PC3 and DU145 cell lines were purchased from the Korean Cell Line Bank (Seoul, Korea). PC3 and DU145 were maintained in RPMI1640 medium (Welgene, Gyeongsan-si, Korea) supplemented with 10% heat-activated fetal bovine serum (Sigma, St. Louis, MO) and 1% penicillin and streptomycin (Thermo, Rockford, IL, USA). Incubator gas tension was maintained at 5% CO2/21%O2 for normoxic conditions and 5% CO2/1%O2 for hypoxic conditions (VS-9000GC; Vision Scientific, Seoul, Korea).
Antibodies and reagents
Culture media and fetal bovine serum was purchased from Sigma-Aldrich (St. Louis, MO, USA). An anti-HIF-1α antibody was generated in rabbits using a bacterially expressed fragment containing amino acids 418–698 of human HIF-1α15. Anti-NDRG3 antiserum was raised from rabbits (New Zealand White) through a commercial facility (AbClon, Seoul, Korea). Rabbits were immunized with a Keyhole limpet hemocyanin (KLH)-conjugated NDRG3 peptide (HSTSSSLGSGESPFSRSVTSNQSDGTQESCESPDVLDRHQTMEVSC). Antibodies against phospho-AKT (S473), total AKT, and Myc-tag were purchased from Cell Signaling (Danvers, MA, USA); anti-Gal4(DBD), anti-β-tubulin, anti-GFP, and HRP-conjugated secondary antibodies from Santa Cruz Biotechnology (Santa Cruz, CA, USA); anti-HA from Roche Diagnostics (Mannheim, Germany); and anti-HIF-2α from Novus Biologicals (Littleton, CO, USA). MK-2206 was purchased from Selleckchem and other chemicals from Sigma-Aldrich.
Preparation of plasmids, short interfering RNAs (siRNAs), and transfection
The cDNA of NDRG3 was cloned by reverse transcription and PCR using Pfu DNA polymerase, and the cDNA was inserted into the MYC-tagged vector by blunt-end ligation. The sequences of siRNA targeting NDRG3 (NM_032013) were 5′-AGAUCAAACCACUUCUAAAUGAUAA-3′ (siNDRG3 #1), 5′-AGAUCAAACCACUUCUAAAUGAUAA-3′ (siNDRG3 #2), and 5′-AGUCAGAUGGAACUCAAGAAUCCTG-3′ (siNDRG3 #3). siRNA targeting HIF-1α was 5′-CAAAGUUAAAGCAUCAGGUUCCUUCUU-3′ (siHIF-1α), siRNA targeting HIF-2α was 5′-GGGUUACUGACGUGTAAAUGCTGGU-3′ (siHIF-2α), and the non-targeting siRNA sequence was 5′-AUGAACGUGAAUUGCUCAATT-3′. Gal4-CAD (HIF-1α a.a. 776–826), Gal4-CAD-N803A, Gal4-NAD (HIF-1α a.a 498–603), and VP16-p300 CH1 plasmids were constructed, as previously described16. For transient gene silencing or protein expression, 40% confluent cells were transfected with plasmids or siRNAs using Lipofectamine 3000 or Lipofectamine RNA iMAX, respectively. The transfected cells were stabilized for 48 h before being used in experiments.
Reporter gene construction and luciferase assay
The luciferase reporter genes containing the hypoxia response element (HRE) of the erythropoietin enhancer or the mutated HRE were kindly provided by Dr. Eric Huang (University of Utah). To determine the cap-dependent translation of HIF-1α, the HIF-1α 5′UTR (1–284) segment was cloned using reverse trancriptase polymerase chain reaction (RT-PCR) and was inserted between the thymidine kinase promoter and luciferase in the GL3 plasmid, as previously described17. PC3 or DU145 cells were co-transfected with 1 μg of the reporter plasmid, 1 μg of the CMV-β-galactosidase plasmid, 40 nM NDRG3 siRNAs, or 1 μg of the NDRG3 plasmid. After stabilization for 48 h, the cells were incubated in 1% O2 or 21% O2 for 24 h, and luciferase activities in the cell lysates were measured using a Lumat LB9507 luminometer (Berthold Technologies, Bad Wildbad, Germany). The specific reporter activity was calculated by dividing luciferase activity by β-galactosidase activity.
Gal4 reporter and mammalian two-hybrid assays
To evaluate HIF-1α CAD activity, PC3 and DU145 cells were co-transfected with 100 ng of Gal4-CAD (or CAD N803A) plasmid, 100 ng of Gal4-Luc plasmid, 500 ng of β-galactosidase plasmid, 40 nM NDRG3 siRNA, or 1 μg of MYC-NDRG3 plasmid using Lipofectamine 3000. For mammalian two-hybrid assays, PC3 and DU145 cells were co-transfected with 100 ng of Gal4-CAD plasmid, 100 ng of Gal4-Luc plasmid, 500 ng of CH1-VP16 plasmid, 500 ng of CMV-β-galactosidase plasmid, 40 nM NDRG3 siRNA, or 1 μg of MYC-NDRG3 plasmid using Lipofectamine 3000. After stabilization for 48 h, the cells were incubated in 1% O2 or 21% O2 for 24 h, and luciferase activities in the cell lysates were measured using a Lumat LB9507 luminometer (Berthold Technologies, Bad Wildbad, Germany). The specific reporter activity was calculated by dividing luciferase activity by β-galactosidase activity.
Immunoblotting and immunoprecipitation
Cell lysates were separated on sodium dodecyl sulfate (SDS)-polyacrylamide gels and transferred to Immobilon-P membranes (Millipore, Bedford, MA, USA). The membranes were blocked with a Tris/saline solution containing 5% skim milk and 0.1% Tween-20 for 1 h and incubated with a primary antibody overnight at 4 °C. Membranes were incubated with a horseradish peroxidase-conjugated secondary antibody for 1 h and visualized using an ECL kit (Thermo, Rockford, IL, USA). To analyze protein interactions, cell lysates were incubated with anti-HA or anti-MYC antibody for 4 h at 4 °C, and the immune complexes were precipitated with protein A/G beads (Santa Cruz, CA, USA). Precipitated proteins were eluted in a denaturing 2× SDS sample buffer, loaded on SDS-PAGE, and immunoblotted.
Cell migration and invasion assays
Transfected PC3 or DU145 cells were cultured in 6.5-mm transwell inserts with an 8 μm pore size, coated with either collagen I or 0.5 mg/μL of growth factor-reduced Matrigel, purchased from Corning Life Science (Acton, MA, USA). Cells were seeded into the upper chambers in 100 μL of fetal bovine serum (FBS)-free medium, while 10% FBS-containing medium was placed in the lower chambers as a chemo-attractant, and incubated in either 1% O2 or 21% O2 for 24 h. Cells on the lower surface of the transwell inserts were fixed in 4% formaldehyde and stained with hematoxylin and eosin, and four high-power independent fields of each membrane were counted.
Quantitative RT-PCR
Total RNA was isolated using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA), and cDNA synthesis was performed in a reaction mixture (Promega, Madison, WI, USA) containing M-MLV Reverse Transcriptase, RNase inhibitor, dNTPs, and random primers at 46 °C for 1 h. Quantitative real-time PCR on 96-well optical plates was performed in the qPCR Mastermix (Enzynomics, Daejeon, Korea), and fluorescence emitting from a dye-DNA complex was monitored in a CFX Connect Real-Time Cycler (BIO-RAD, Hercules, CA). The mRNA values of targeted genes were calculated relative to GAPDH expression. All reactions were performed in triplicate. The nucleotide sequences of PCR primers are summarized in Table S1.
Chromatin immunoprecipitation
Cells were fixed with 37% formaldehyde at 37 °C for 10 min and then treated with 150 mM glycine. Fixed cells were lysed with 0.5% NP-40 and centrifuged at 800 × g at 4 °C for 10 min to collect the crude nuclear fraction. The nuclear pellet was incubated with 1% SDS and was subjected to sonication to shear genomic DNA into 300–500 bp fragments. Soluble chromatin complexes were immunoprecipitated with IgG, anti-HIF-1α, or anti-p300 antibody overnight at 4 °C. Immune complexes were precipitated with protein A/G beads pre-blocked by salmon sperm DNA at 4 °C for 4 h. The beads were sequentially washed with a low salt buffer, a high salt buffer, LiCl wash buffer, and TE buffer. The immunoprecipitates were eluted in a chromatin immunoprecipitation (ChIP) direct elution buffer at 65 °C for 30 min. DNA was isolated by phenol–chloroform–isoamyl alcohol (25:24:1) and precipitated with ethanol and glycogen. The extracted DNA was resolved in nuclease-free water and analyzed by real-time PCR (95/55/72 °C, 30 s at each phase).
Immunohistochemistry in human prostate cancer tissues
Human prostate cancer tissue microarrays were purchased from SuperBioChips Lab (Seoul, South Korea). Tumor staging was defined according to the AJCC cancer staging manual (7th edition). Clinical information on prostate cancer patients is summarized in Table S2. Paraffin-fixed tissue slides were incubated in a 60 °C oven for 1 h to remove paraffin and were autoclaved in antigen retrieval solution. After treatment with 3% H2O2, tissue sections were incubated with primary antibodies (anti-NDRG3 and anti-HIF-1α) overnight at 4 °C, followed by biotinylated secondary antibodies for 1 h at 25 °C. The immune complexes were visualized using a Vectastain ABC kit (Vector Laboratories, Burlingame, CA, USA) and counterstained with hematoxylin for 10 min. Protein expression levels were analyzed by counting positively stained cells in four independent high-power fields on each slide.
Informatics analysis
The normalized data set GSE6919 for prostate cancer was imported from Gene Expression Omnibus (GEO), in which the total patients (n = 167) were categorized into four groups: normal prostate tissue free of any pathological alterations (n = 17), normal prostate tissue adjacent to tumor (n = 59), primary prostate tumor (n = 66), and metastatic prostate tumor (n = 25). For mRNA expression analysis, normalized values of 57107_at (corresponding to NDRG3) in each group were compared between three groups: normal prostate tissue adjacent to tumor, primary prostate tumor, and metastatic prostate tumor. The group that consisted of ‘normal prostate tissue free of any pathological alterations’ (n = 18) was excluded from mRNA analysis. For gene set enrichment analysis (GSEA), a formatted GCT file was used as the input for the GSEA algorithm v2.0 (available from: http://www.broadinstitute.org/gsea). For grouping the GSE6919 data set, the values of the 57107_at (corresponding to NDRG3) were used as the criteria standard for the low expression and high expression groups. The phenotype and the default parameters were used with the results that the Pearson correlation was computed from to rank the genes.
Statistics
All data were analyzed using Microsoft Excel 2013 software or GraphPad Prism 5 software, and the results are expressed as the means and standard deviations. We used unpaired, two-sided Student’s t-test to compare protein levels, luciferase activities, and cell numbers. Differences were considered statistically significant when P-values were less than 0.05. Protein or mRNA expression correlations were analyzed using a Spearman’s P statistics. Survival rate analysis was performed by drawing curves and calculating the log-rank P test using the Kaplan–Meier method.
Results
NDRG3 and HIF-α reciprocally regulate each other at the protein level
We first examined the temporal patterns of NDRG3 and HIF-1/2α expression during hypoxia. As has been previously reported18, HIF-1/2α levels in prostate cancer cells increased as early as 4 h after hypoxia and subsided after 16 h. In contrast, NDRG3 started to be induced 16 h after hypoxia and its expression gradually increased with increasing time of hypoxia (Fig. 1a). We noted that HIF-1/2α expression preceded NDRG3 expression and that NDRG3 expression preceded a gradual decline of HIF-1/2α. Given the kinetic patterns of HIF-1/2α and NDRG3 expression, we suspected a reciprocal regulation between them. When HIF-1α or HIF-2α was knocked down in prostate cancer cells, the hypoxic induction of NDRG3 was diminished (Fig. 1b) whereas the NDRG3 mRNA level was not altered (Supplementary Figure S1a), suggesting that HIF-1/2α do not act as the transcription factors for the NDRG3 gene. Indeed, the HIF-dependent expression of NDRG3 has been demonstrated by a recent report14. Lee et al. suggested that HIFs promote lactate production by upregulating a series of glycolytic enzymes and the accumulated lactate potentiates the hypoxic stabilization of NDRG3 by inhibiting PHD docking to NDRG3. Alternatively, we found that the hypoxic induction of HIF-1/2α was potentiated by NDRG3 knockdown but weakened by NDRG3 overexpression (Fig. 1c, Supplementary Figure S1b). Such effects of NDRG3 knockdown on HIF-1/2α induction were also observed in glioblastoma U-251 cells, but not in breast cancer MCF7 or lung cancer A549 cells (Supplementary Figure S1c). The NDRG3 inhibition of HIF-1/2α expression seems to occur in a cell context-dependent manner. As expected, the mRNA levels of HIF-downstream genes PDK1 and BNIP3 in prostate cancer cells were significantly increased under hypoxia by NDRG3 knockdown (Fig. 1d). When hypoxia persists, accordingly, HIFs upregulate NDRG3 and in turn NDRG3 inhibits HIFs. NDRG3 may function to control the HIF-mediated hypoxic response through a negative feedback loop.
a PC3 and DU145 cells were incubated under hypoxia (1% O2) for the indicated times and subjected to western blotting with an anti-HIF-1α or anti-HIF-2α antibody. b PC3 and DU145 cells, which had been transfected with siRNAs silencing HIF-1α or HIF-2α, were incubated under normoxia or hypoxia for 24 h and subjected to western blotting. c PC3 and DU145 cells, which had been transfected with NDRG3-silencing siRNA (siNDRG3 #1), were incubated under normoxia or hypoxia for 24 h and subjected to western blotting. d PC3 and DU145 cells, which had been transfected with NDRG3-silencing siRNA, were incubated under normoxia or hypoxia for 24 h and lysed for RNA extraction. PDK1 and BNIP3 mRNA levels were measured by RT-qPCR. Each bar represents the mean + s.d. (n = 3) and * denotes P < 0.05 between the indicated groups
Cap-dependent translation of HIF-α is reduced by NDRG3
We next proceeded to determine how NDRG3 downregulates HIF-1α and HIF-2α. Because NDRG3 knockdown did not increase HIF-1α or HIF-2α mRNA levels (Fig. 2a), NDRG3 may regulate HIF-1/2α at the post-transcriptional level. Next, we compared the oxygen-dependent degradation rates of HIF-1/2α proteins and found that the stabilities of HIF-1/2α proteins were not significantly altered by NDRG3 knockdown (Fig. 2b, Supplementary Figure S2a). Subsequently, the effect of NDRG3 on HIF-1/2α protein syntheses was examined. The rate of de novo protein synthesis was analyzed in the presence of MG132, which blocks HIF-1/2α degradation. Given that the synthesis rates of HIF-1/2α proteins were enhanced by NDRG3 knockdown (Fig. 2c, Supplementary Figure S2b), NDRG3 likely controls the translation of HIF-1/2α mRNA. We assessed the inhibitory function of NDRG3 on HIF-1α translation using the HIF1A_5′UTR-luciferase reporter, which reflects the cap-dependent translation of HIF-1α. Luciferase activity was largely increased by NDRG3 knockdown in both normoxia and hypoxia (Fig. 2d). This result further supports our notion that NDRG3 negatively regulates de novo synthesis of HIF-1α protein.
a PC3 and DU145 cells, which had been transfected with NDRG3-silencing siRNA, were incubated under hypoxia for 24 h and subjected to RT-qPCR to measure the mRNA levels of HIF-1α and HIF-2α (means + s.d., n = 3). ns denotes “not significantly different.” b The transfected PC3 or DU145 cells were incubated under hypoxia for 8 h, followed by re-oxygenation at 21% O2. HIF-1α and HIF-2α proteins were immunoblotted and their blots were quantified using the ImageJ program. Relative band intensities were normalized to the corresponding β-tubulin intensities. Each symbol represents the mean ± s.d. (n = 3), and the linear regression was plotted using the SigmaPlot program. c The transfected PC3 or DU145 cells were incubated with 15 μM MG132 for the indicated times. Western blot intensities were analyzed using ImageJ, and the relative band intensities were normalized to the corresponding β-tubulin intensities. Each symbol represents the mean ± s.d. (n = 3). * denotes P < 0.05 versus the siCtrl group at the corresponding time. d PC3 or DU145 cells were co-transfected with the TK-HIF1A 5′UTR-luciferase plasmid, the CMV-β-galactosidase plasmid, and siCtrl or siNDRG3. After stabilization for 24 h, the transfected cells were incubated under normoxia or hypoxia for 24 h. Luciferase activities were measured and normalized to β-galactosidase activities. Each bar represents the mean + s.d. (n = 3) and * denotes P < 0.05 between the indicated groups
NDRG3 inhibits the AKT-dependent syntheses of HIF-1/2α proteins
The cap-dependent translation of HIF-1/2α in prostate cancer cells is promoted through the PI3K–AKT–mTOR pathway12,19,20,21. Therefore, we tested whether NDRG3 controls this pathway and found that NDRG3 negatively regulates AKT phosphorylation in prostate cancer cells (Fig. 3a, Supplementary Figure S3a). Furthermore, HIF-1α and HIF-2α syntheses induced by NDRG3 knockdown were abolished by an AKT inhibitor MK-2206 (Fig. 3b, Supplementary Figure S3b). As shown in the HIF1A_5′UTR-lucifease reporter system, the cap-dependent translation of HIF-1α was significantly decreased by MK-2206 (Fig. 3c). Taken together, NDRG3 might function to control HIF-1/2α expression by inhibiting the AKT-dependent translation of their mRNAs.
a Phospho-AKT (S473) and total AKT levels were analyzed in PC3 and DU145 cells that were transfected with siNDRG3 and incubated under hypoxia for 24 h. b The transfected PC3 or DU145 cells were pre-treated with 1 μM MK-2206 for 16 h and incubated with 15 μM MG132 for the indicated times. Western blot intensities were analyzed using ImageJ, and the relative band intensities were normalized to the corresponding β-tubulin intensities. Each symbol represents the mean ± s.d. (n = 3). c The TK-HIF1A 5′UTR-luciferase reporter and the CMV-β-galactosidase plasmids were co-transfected with siCtrl or siNDRG3 into PC3 and DU145 cells. After stabilization for 24 h, the cells were incubated with DMSO or MK-2206 under normoxia or hypoxia for 24 h. Luciferase activities (means + s.d., n = 3) were normalized to β-galactosidase activities. * denotes P < 0.05 between the indicated groups
NDRG3 inhibits the HIF-driven transcription of the HRE promoter
To examine whether NDRG3 controls HIF-driven gene expression under hypoxia, an HRE-luciferase reporter plasmid, which contains the HRE from the erythropoietin enhancer region, was utilized. The hypoxic enhancement of the reporter activity was increased by NDRG3 knockdown (Fig. 4a) but attenuated by NDRG3 overexpression (Fig. 4b). As the mutated HRE reporter activity was constant regardless of NDRG3 expression, NDRG3 controls the reporter activity by specifically regulating HIF-1/2. To examine if NDRG3 knockdown enhances HIF-driven transcription through AKT signaling, we analyzed the reporter activity in the presence of MK-2206. The NDRG3 knockdown-induced increase of the luciferase activity was significantly, but not fully, diminished by MK-2206 (Fig. 4c). This result suggests that NDRG3 inhibition of HIF-driven transcription is partially attributed to the reduction in AKT-dependent synthesis of HIF-1/2α proteins. In addition to AKT inhibition, however, another mechanism may contribute to the functional inhibition of HIF-1/2α by NDRG3.
a PC3 and DU145 cells were co-transfected with the HRE- or mutated HRE-luciferase plasmid, the CMV-β-galactosidase plasmid, and siCtrl or siNDRG3, and incubated under normoxia or hypoxia for 24 h. b Cells were co-transfected with the reporter, the CMV-β-galactosidase, and pcDNA or MYC-NDRG3 plasmids, and incubated under normoxia or hypoxia for 24 h. c Cells were co-transfected with the HRE reporter plasmid, the β-galactosidase plasmid, and siCtrl or siNDRG3. The cells were treated with DMSO or MK-2206 and incubated under normoxia or hypoxia for 24 h. Luciferase activities (means + s.d., n = 3) were normalized to β-galactosidase activities. * denotes P < 0.05 between the indicated groups
NDRG3 functionally inhibits HIF-1 by interfering with p300 binding to HIF-1α
Next, we tested whether NDRG3 controls the hypoxic activation of HIF-1α_CAD (the transactivation domain at the C-terminus) using the Gal4 reporter system, which contains the Gal4 DNA-binding domain/HIF-1α_CAD fusion gene and the Gal4 promoter-luciferase fusion gene. Because the Gal4-CAD fusion protein is constantly expressed regardless of the oxygen tension (Figure S4a), this reporter system can reflect the transcriptional activity of HIF-1α irrespective of HIF-1α stability. Aside from the change in protein synthesis, the transcriptional activity of HIF-1α was also elevated by NDRG3 knockdown but was repressed by NDRG3 overexpression (Fig. 5a). HIF-1α_CAD is oxygen-dependently inactivated by FIH, which enzymatically hydroxylates the N803 residue of CAD10. To examine whether NDRG3 regulates this FIH activity, we measured the activity of the Gal4-CAD_N803A mutant, which is not regulated by FIH. Unexpectedly, NDRG3 controlled the CAD activity irrespective of the N803 hydroxylation by FIH (Fig. 5b). Next, we assessed the interaction between HIF-1α_CAD and the CH1 domain of p300, which is an essential process for HIF-driven transcription. For this analysis, we used the mammalian two-hybrid system, which contains Gal4 promoter-luciferase, Gal4-CAD, and VP16-CH1 plasmids. Consequently, the CAD–CH1 interaction was markedly enhanced by NDRG3 knockdown but almost completely inhibited by NDRG3 overexpression (Fig. 5c), strongly indicating that NDRG3 interferes with p300 binding to HIF-1α. To confirm this effect of NDRG3, co-immunoprecipitation was performed. GFP-HIF-1α and HA-p300 were co-expressed in PC3 cells, and the cell lysates were immunoprecipitated with anti-HA beads. With increasing expression of NDRG3, the interaction between HIF-1α and p300 was reduced (Fig. 5d). These findings suggest that NDRG3 represses HIF-driven transcription by dissociating p300 from HIF-1α. In addition, NDRG3 was silenced in PC3 cells and then ChIP-qPCR was performed to detect HIF-1α or p300 binding to the BNIP3 promoter region. NDRG3 knockdown greatly increased the binding between the BNIP3 promoter and HIF-1α or p300, further suggesting that HIF-1α transcriptional activity is repressed by NDRG3 (Supplementary Figure S4b).
a, b The MYC-NDRG3 plasmid or siNDRG3 was co-transfected with the Gal4 promoter-Luc reporter plasmid and the Gal4/DBD-HIF-1α_CAD plasmid or the Gal4/DBD-HIF-1α_CAD N803A plasmid into PC3 and DU145 cells. Cells were incubated under normoxia or hypoxia for 24 h. c The MYC-NDRG3 plasmid or siNDRG3 was co-transfected with Gal4 promoter-Luc reporter, Gal4/DBD-HIF-1α_CAD, and p300_CH1-VP16 plasmids. Cells were incubated under normoxia or hypoxia for 24 h. Luciferase activities (means + s.d., n = 3) were normalized to β-galactosidase activities and presented as relative values to the normoxic control. * denotes P < 0.05 between the indicated groups. d PC3 cells were co-transfected with GFP-HIF-1α, HA-p300, and MYC-NDRG3 (0.5 or 2 μg). Proteins in cell lysates were immunoprecipitated with an anti-HA antibody and immunoblotted using the indicated antibodies. The experiments were performed three times
NDRG3 inhibits the hypoxia-induced migration and invasion of prostate cancer cells
Hypoxia-induced cancer metastasis is regarded as one of the most important roles of HIF-1α. HIF-1α can induce cancer metastasis by expressing EMT-inducing genes, such as Twist and Snail22,23,24. Therefore, we proceeded to study the role of NDRG3 in cell migration and invasion in prostate cancer. We first examined the effects of NDRG3 on EMT marker expression and found that the protein level of an epithelial marker E-cadherin was decreased by NDRG3 knockdown while the levels of mesenchymal markers, such as N-cadherin, α-SMA, and Twist, were increased (Supplementary Figure S5). In a transwell migration assay, cell migration under hypoxia was facilitated by NDRG3 knockdown but was attenuated by NDRG3 overexpression in PC3 cells (Fig. 6a) and DU145 cells (Fig. 6b). Furthermore, cell invasion was also negatively regulated by NDRG3 in PC3 cells (Fig. 6c) and DU145 cells (Fig. 6d).
PC3 (a, c) or DU145 (b, d) cells were transfected with siNDRG3 or MYC-NDRG3, and 1 × 104 cells were seeded in a transwell culture plate. The upper chambers were filled with serum-free media and the lower chambers with 10% FBS-containing media. Migration and invasion analyses were performed using uncoated and Matrigel-coated interface membranes, respectively. After cells were incubated under normoxia or hypoxia for 24 h, the membranes were fixed in 4% PFA and subjected to H&E staining. Representative images of each group are shown on the left, and the numbers (means + s.d., n = 3) of migrated or invaded cells are shown as bar graphs on the right. * denotes P < 0.05
NDRG3 expression inversely correlates with metastasis and HIF-1α expression in human prostate cancer
To investigate the role of NDRG3 in prostate cancer progression, NDRG3 mRNA levels in prostate cancer tissues were analyzed using the NCBI GEO data set GSE6919. NDRG3 mRNA levels were significantly lower in metastatic prostate cancer tissues than in primary cancer tissues or normal tissues (Fig. 7a). Based on the median value (961.25) of NDRG3 mRNA (Fig. 7b), prostate cancer tissues were divided into two groups, NDRG3_Low and NDRG3_High. We performed GSEA using the HIF-1 downstream gene set HIF1_TFPATHWAY and found that HIF-1 downstream genes are significantly enriched in the NDRG3_Low group (Fig. 7c, Supplementary Figures S6a–c, Supplementary Tables S3–6). These informatics analyses prompted us to assess the NDRG3 protein levels in prostate cancer tissues obtained from patients. Immunohistochemical analyses showed that NDRG3 expression in prostate cancer was gradually reduced with increasing Gleason score (Fig. 7d). In contrast, HIF-1α expression was increased in cancer with a high Gleason score, which is consistent with other reports25,26. Furthermore, patients in the NDRG3_Low group tended to have lower disease-free survival rates than those in the NDRG3_High group while patients in the HIF-1α_Low group showed a higher survival rate (Fig. 7e). Pearson correlation analysis also demonstrated a negative correlation between NDRG3 and HIF-1α protein expression in prostate cancer tissues (Fig. 7f), further supporting the notion that NDRG3 is a negative regulator of HIF-1α. Taken together, we propose that NDRG3 plays an anti-metastatic role in prostate cancer by antagonizing the HIF signaling pathway.
a Relative NDRG3 mRNA expression in normal adjacent to tumor tissues (NAT, n = 59), primary tumor tissues (PT, n = 66), and metastatic tumor tissues (MT, n = 25), which were obtained from GEO data set GSE6919. b The GSE6919 data set is divided into NDRG3_Low and NDRG3_High groups according to the median value (961.25) of the NDRG3 mRNA level. c The GSEA enrichment plot for the PID_HIF1_TFPATHWAY gene set in the NDRG3_Low group. d IHC analyses of NDRG3 and HIF-1α protein expressions in a human prostate adenocarcinoma tissue microarray. Representative images are shown on the left and dot plots of the immunostaining scores are shown on the right. The scale bar represents 50 μm. e Kaplan–Meier analysis for the association of NDRG3 or HIF-1α expression and the tumor-free survival rates in the human prostate adenocarcinoma tissue microarray. The Low and High groups were categorized according to the median values of the protein expressions. P-value was calculated by log-rank test. f Pearson correlation analysis shows a negative correlation between NDRG3 and HIF-1α expression in the tissue microarray. R value indicates the Pearson correlation coefficient value. The horizontal lines in all dot plots represent the means ± s.e.m. and * denotes P < 0.05 between the indicated groups
Discussion
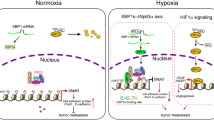
Despite the numerous benefits of HIF-1α in hypoxic responses, a prolonged expression of HIF-1α eventually threatens cell survival during long-term hypoxia because pro-apoptotic genes are expressed by HIF-1α18,27,28. Therefore, it is reasonable that HIF-1α expression is robustly induced during acute hypoxia but gradually subsides in later phases of hypoxia29,30. We also observed the downregulation of HIF-1α proteins in PC3 and DU145 cells after 16 h of hypoxia. Despite many efforts, the mechanism underlying the temporal readjustment of HIF-1α expression has not been clearly elucidated. Based on our results, we propose a new hypothesis for this mechanism. While hypoxia persists, HIF-1α expression is followed by NDRG3 expression because NDRG3 stabilization requires lactate accumulation through HIF-1-induced glycolytic enzymes. After NDRG3 accumulates in cells, it not only downregulates HIF-1α protein at the translational level but also represses the transcriptional activity of HIF-1α at the epigenetic level. By doing so, NDRG3 controls HIF-mediated cell migration and metastasis in prostate cancer. The proposed role of NDRG3 as a feedback regulator of HIF-1 is summarized in Fig. 8.
To date, several hypotheses have been proposed to explain how HIF-1α expression subsides in long-term hypoxia. For example, HSP70 recruits the chaperone-dependent E3 ubiquitin ligase CHIP (carboxyl terminus of Hsc70-interacting protein) to HIF-1α, leading to HIF-1α ubiquitination and degradation during prolonged hypoxia31. REST (repressor element 1-silencing transcription factor) or PRDX2/4 (peroxiredoxin 2 and 4) targets and inactivates the promoter of the HIF1A gene, which downregulates HIF-1α at the transcriptional level in prolonged hypoxia32,33. PHD2/3, whose expression is induced under hypoxia, facilitates the prolyl hydroxylation of HIF-1α even under hypoxia, and subsequently downregulates HIF-1α at the post-translational level34,35. In this study, NDRG3 was also identified as a negative feedback regulator of the HIF-mediated hypoxic signaling pathway. Given that HIF-1 signaling is tightly controlled through multiple pathways, HIF-1 might be required for the short-term attention of hypoxia-injured cells but not for the long-term management of cell life.
NDRG3 has been in the spotlight because its regulation is very similar to the oxygen-sensitive degradation of HIF-1α14. However, little is known about the role of NDRG3 in the cellular adaptation to hypoxia. Lee et al. demonstrated that NDRG3 promoted angiogenesis, cell proliferation, and anti-apoptosis by activating the RAF–ERK pathway and that its expression is higher in hepatocellular carcinoma than in normal liver tissue14. According to the report, NDRG3 is considered to be oncogenic in liver cancer.
Previously, NDRG3 was reported as an androgen-regulated gene that presumably increased the malignancy of prostate cancer36. However, our study shows opposing results, as NDRG3 was found to repress HIF-1α activity in prostate cancer cells, and NDRG3 expression was downregulated in advanced prostate cancer tissues. Such conflicting data may result from the fact that our study investigated the role of NDRG3 in hypoxic tumor events. Another recent study showed that NDRG3 was downregulated in advanced breast cancer and that its expression was positively correlated with an increased disease-free survival rate37. This report supports the tumor-suppressive function of NDRG3 in breast cancer. Given such conflicting reports on NDRG3 function, NDRG3 may play differential roles in cancer progression depending on cell context. The cancer type-dependent roles of NDRG3 should be further investigated.
In conclusion, our results revealed a novel mechanism of HIF-1α autoregulation through the cross-talk with hypoxia-inducible NDRG3. NDRG3 is induced by the HIF-1 signaling pathway, which in turn blocks HIF-1α synthesis and also represses the transcriptional activity of HIF-1α. This study also provides new insight into the tumor-suppressive role of NDRG3 in prostate cancer. NDRG3 may be a potential target in developing an HIF-targeting anticancer strategy.
References
Semenza, G. L. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 7, 345–350 (2001).
Semenza, G. L. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15, 551–578 (1999).
Wang, G. L., Jiang, B. H., Rue, E. A. & Semenza, G. L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 92, 5510–5514 (1995).
Semenza, G. L. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda). 19, 176–182 (2004).
Kaelin, W. G. Jr & Ratcliffe, P. J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 (2008).
Huang, L. E., Gu, J., Schau, M. & Bunn, H. F. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA 95, 7987–7992 (1998).
Jaakkola, P. et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001).
Schofield, C. J. & Ratcliffe, P. J. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell. Biol. 5, 343–354 (2004).
Lando, D. et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16, 1466–1471 (2002).
Lando, D., Peet, D. J., Whelan, D. A., Gorman, J. J. & Whitelaw, M. L. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science 295, 858–861 (2002).
Shukla, S. et al. Activation of PI3K-Akt signaling pathway promotes prostate cancer cell invasion. Int. J. Cancer 121, 1424–1432 (2007).
Zhong, H. et al. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 60, 1541–1545 (2000).
Lee, G. Y., Chun, Y. S., Shin, H. W. & Park, J. W. Potential role of the N-MYC downstream-regulated gene family in reprogramming cancer metabolism under hypoxia. Oncotarget 7, 57442–57451 (2016).
Lee, D. C. et al. A lactate-induced response to hypoxia. Cell 161, 595–609 (2015).
Chun, Y. S. et al. Zinc induces the accumulation of hypoxia-inducible factor (HIF)-1alpha, but inhibits the nuclear translocation of HIF-1beta, causing HIF-1 inactivation. Biochem. Biophys. Res. Commun. 268, 652–656 (2000).
To, K. K. & Huang, L. E. Suppression of hypoxia-inducible factor 1alpha (HIF-1alpha) transcriptional activity by the HIF prolyl hydroxylase EGLN1. J. Biol. Chem. 280, 38102–38107 (2005).
Shin, H. W., Cho, C. H., Kim, T. Y. & Park, J. W. Sunitinib deregulates tumor adaptation to hypoxia by inhibiting HIF-1alpha synthesis in HT-29 colon cancer cells. Biochem. Biophys. Res. Commun. 398, 205–211 (2010).
Ginouves, A., Ilc, K., Macias, N., Pouyssegur, J. & Berra, E. PHDs overactivation during chronic hypoxia “desensitizes” HIFalpha and protects cells from necrosis. Proc. Natl. Acad. Sci. USA 105, 4745–4750 (2008).
Block, K. et al. NAD(P)H oxidases regulate HIF-2alpha protein expression. J. Biol. Chem. 282, 8019–8026 (2007).
Khatua, S. et al. Overexpression of the EGFR/FKBP12/HIF-2alpha pathway identified in childhood astrocytomas by angiogenesis gene profiling. Cancer Res. 63, 1865–1870 (2003).
Semenza, G. L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3, 721–732 (2003).
Yang, M. H. & Wu, K. J. TWIST activation by hypoxia inducible factor-1 (HIF-1): implications in metastasis and development. Cell Cycle 7, 2090–2096 (2008).
Imai, T. et al. Hypoxia attenuates the expression of E-cadherin via up-regulation of SNAIL in ovarian carcinoma cells. Am. J. Pathol. 163, 1437–1447 (2003).
Yang, M. H. et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat. Cell Biol. 10, 295–305 (2008).
Boddy, J. L. et al. The androgen receptor is significantly associated with vascular endothelial growth factor and hypoxia sensing via hypoxia-inducible factors HIF-1a, HIF-2a, and the prolyl hydroxylases in human prostate cancer. Clin. Cancer Res. 11, 7658–7663 (2005).
Zhong, H. et al. Increased expression of hypoxia inducible factor-1alpha in rat and human prostate cancer. Cancer Res. 58, 5280–5284 (1998).
Carmeliet, P. et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394, 485–490 (1998).
Greijer, A. E. & van der Wall, E. The role of hypoxia inducible factor 1 (HIF-1) in hypoxia induced apoptosis. J. Clin. Pathol. 57, 1009–1014 (2004).
Henze, A. T. et al. Prolyl hydroxylases 2 and 3 act in gliomas as protective negative feedback regulators of hypoxia-inducible factors. Cancer Res. 70, 357–366 (2010).
Uchida, T. et al. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression in lung epithelial cells: implication of natural antisense HIF-1alpha. J. Biol. Chem. 279, 14871–14878 (2004).
Luo, W. et al. Hsp70 and CHIP selectively mediate ubiquitination and degradation of hypoxia-inducible factor (HIF)-1alpha but Not HIF-2alpha. J. Biol. Chem. 285, 3651–3663 (2010).
Cavadas, M. A. et al. REST mediates resolution of HIF-dependent gene expression in prolonged hypoxia. Sci. Rep. 5, 17851 (2015).
Luo, W. et al. PRDX2 and PRDX4 are negative regulators of hypoxia-inducible factors under conditions of prolonged hypoxia. Oncotarget 7, 6379–6397 (2016).
Minamishima, Y. A. et al. A feedback loop involving the Phd3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol. Cell Biol. 29, 5729–5741 (2009).
Stiehl, D. P. et al. Increased prolyl 4-hydroxylase domain proteins compensate for decreased oxygen levels. Evidence for an autoregulatory oxygen-sensing system. J. Biol. Chem. 281, 23482–23491 (2006).
Wang, W. et al. NDRG3 is an androgen regulated and prostate enriched gene that promotes in vitro and in vivo prostate cancer cell growth. Int. J. Cancer 124, 521–530 (2009).
Estiar, M. A. et al. Clinical significance of NDRG3 in patients with breast cancer. Future Oncol. 13, 961–969 (2017).
Acknowledgements
The authors thank Dr. Woo Ho Kim (Seoul National University) for kindly supplying prostate cancer arrays and Dr. Eric Huang (University of Utah) for kindly providing HRE-Luc, mHRE-Luc, GAL4-Luc, and GAL4-CAD plasmids. This work was supported by grants from the National Research Foundation of Korea (2017015015 and 2017048432).
Author information
Authors and Affiliations
Contributions
G.Y.L. and J.-W.P. designed the study and wrote the manuscript. J.-W.P. and Y.-S.C. cloned cDNAs and constructed expression plasmids. G.Y.L. performed cell-based experiments and immunohistochemistry on the cancer tissue array. G.Y.L., S.-H.S., and H.-W.S. analyzed informatics.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, and provide a link to the Creative Commons license. You do not have permission under this license to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lee, G.Y., Shin, SH., Shin, HW. et al. NDRG3 lowers the metastatic potential in prostate cancer as a feedback controller of hypoxia-inducible factors. Exp Mol Med 50, 1–13 (2018). https://doi.org/10.1038/s12276-018-0089-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-018-0089-y
This article is cited by
-
A SUMOylation-dependent HIF-1α/CLDN6 negative feedback mitigates hypoxia-induced breast cancer metastasis
Journal of Experimental & Clinical Cancer Research (2020)
-
Oxygen sensor FIH inhibits HACE1-dependent ubiquitination of Rac1 to enhance metastatic potential in breast cancer cells
Oncogene (2019)
-
PIN1 transcript variant 2 acts as a long non-coding RNA that controls the HIF-1-driven hypoxic response
Scientific Reports (2019)