Abstract
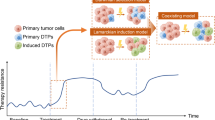
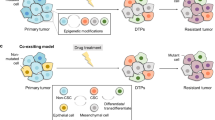
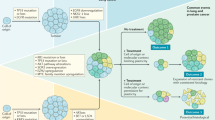
The success of targeted therapies in cancer treatment has been impeded by various mechanisms of resistance. Besides the acquisition of resistance-conferring genetic mutations, reversible mechanisms that lead to drug tolerance have emerged. Plasticity in tumour cells drives their transformation towards a phenotypic state that no longer depends on the drug-targeted pathway. These drug-refractory cells constitute a pool of slow-cycling cells that can either regain drug sensitivity upon treatment discontinuation or acquire permanent resistance to therapy and drive relapse. In the past few years, cell plasticity has emerged as a mode of targeted therapy evasion in various cancers, ranging from prostate and lung adenocarcinoma to melanoma and basal cell carcinoma. Our understanding of the mechanisms that control this phenotypic switch has also expanded, revealing the crucial role of reprogramming factors and chromatin remodelling. Further deciphering the molecular basis of tumour cell plasticity has the potential to contribute to new therapeutic strategies which, combined with existing anticancer treatments, could lead to deeper and longer-lasting clinical responses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hanahan, D. Rethinking the war on cancer. Lancet 383, 558–563 (2014).
Redmond, K. M., Wilson, T. R., Johnston, P. G. & Longley, D. B. Resistance mechanisms to cancer chemotherapy. Front. Biosci. 13, 5138–5154 (2008).
Roesch, A. Tumor heterogeneity and plasticity as elusive drivers for resistance to MAPK pathway inhibition in melanoma. Oncogene 34, 2951–2957 (2015).
Hata, A. N. et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med. 22, 262–269 (2016). This paper shows that cancer cells with resistance-conferring mutations can either pre-exist or evolve from drug-tolerant cells.
Salgia, R. & Kulkarni, P. The genetic/non-genetic duality of drug 'resistance' in cancer. Trends Cancer 4, 110–118 (2018).
Sharma, S. V. et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010). This is the first study to identify the persistence of drug-tolerant cells upon treatment with various anticancer agents in vitro.
Oser, M. G., Niederst, M. J., Sequist, L. V. & Engelman, J. A. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 16, e165–e172 (2015). This is an insightful review of SCLC transformation in lung adenocarcinoma.
Biehs, B. et al. A cell identity switch allows residual BCC to survive Hedgehog pathway inhibition. Nature 562, 429–433 (2018).
Sanchez-Danes, A. et al. A slow-cycling LGR5 tumour population mediates basal cell carcinoma relapse after therapy. Nature 562, 434–438 (2018). This study and reference 8 discuss the mechanisms underlying the survival of tumour cells upon vismodegib treatment in BCC.
Davies, A. H., Beltran, H. & Zoubeidi, A. Cellular plasticity and the neuroendocrine phenotype in prostate cancer. Nat. Rev. Urol. 15, 271–286 (2018). This is a comprehensive review on neuroendocrine differentiation in prostate cancer.
Su, Y. et al. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc. Natl Acad. Sci. USA 114, 13679–13684 (2017).
Rambow, F. et al. Toward minimal residual disease-directed therapy in melanoma. Cell 174, 843–855.e819 (2018). This study and reference 11 were the first to show that new cell states can emerge upon drug treatment and coexist in residual melanoma cells.
Vallette, F. M. et al. Dormant, quiescent, tolerant and persister cells: four synonyms for the same target in cancer diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Biochem. Pharmacol. 7, 10690 (2018).
Hammerlindl, H. & Schaider, H. Tumor cell-intrinsic phenotypic plasticity facilitates adaptive cellular reprogramming driving acquired drug resistance. J. Cell Commun. Signal 12, 133–141 (2018). This review analyses phenotypic plasticity upon targeted therapies and the emergence of slow-cycling drug-tolerant cells in vitro.
Luskin, M. R., Murakami, M. A., Manalis, S. R. & Weinstock, D. M. Targeting minimal residual disease: a path to cure? Nat. Rev. Cancer 18, 255–263 (2018).
Ramirez, M. et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat. Commun. 7, 10690 (2016).
Roesch, A. et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 23, 811–825 (2013).
Trumpp, A. & Wiestler, O. D. Mechanisms of disease: cancer stem cells–targeting the evil twin. Nat. Clin. Prac. Oncol. 5, 337–347 (2008).
Glasspool, R. M., Teodoridis, J. M. & Brown, R. Epigenetics as a mechanism driving polygenic clinical drug resistance. Br. J. Cancer 94, 1087–1092 (2006).
Shaffer, S. M. et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017). This paper shows transient expression of resistance markers in rare cells that are more likely to survive drug treatment.
Kurata, T. et al. Effect of re-treatment with gefitinib ('Iressa', ZD1839) after acquisition of resistance. Ann. Oncol. 15, 173–174 (2004).
Yano, S. et al. Retreatment of lung adenocarcinoma patients with gefitinib who had experienced favorable results from their initial treatment with this selective epidermal growth factor receptor inhibitor: a report of three cases. Oncol. Res. 15, 107–111 (2005). This study and reference 21 discuss clinical cases of response to EGFR-TKI retreatment after a drug holiday.
Tang, J. Y. et al. Inhibition of the hedgehog pathway in patients with basal-cell nevus syndrome: final results from the multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 17, 1720–1731 (2016).
Tata, P. R. & Rajagopal, J. Cellular plasticity: 1712 to the present day. Curr. Opin. Cell Biol. 43, 46–54 (2016).
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017).
Sequist, L. V. et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci, Transl Med. 3, 75ra26 (2011).
Shibue, T. & Weinberg, R. A. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 14, 611–629 (2017). This is an insightful review about the role of EMT in resistance to therapy.
Fischer, K. R. et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 (2015).
Zheng, X. et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530 (2015).
Yauch, R. L. et al. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin. Cancer Res. 11, 8686–8698 (2005).
Zhang, Z. et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat. Genet. 44, 852–860 (2012).
Rho, J. K. et al. Epithelial to mesenchymal transition derived from repeated exposure to gefitinib determines the sensitivity to EGFR inhibitors in A549, a non-small cell lung cancer cell line. Lung Cancer 63, 219–226 (2009).
Byers, L. A. et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin. Cancer Res. 19, 279–290 (2013).
Weng, C. H. et al. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene 38, 455–468 (2019).
Chung, J. H. et al. Clinical and molecular evidences of epithelial to mesenchymal transition in acquired resistance to EGFR-TKIs. Lung Cancer 73, 176–182 (2011).
Wu, W. S. et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell 123, 641–653 (2005).
Saxena, M., Stephens, M. A., Pathak, H. & Rangarajan, A. Transcription factors that mediate epithelial-mesenchymal transition lead to multidrug resistance by upregulating ABC transporters. Cell Death Dis. 2, e179 (2011).
Oshimori, N., Oristian, D. & Fuchs, E. TGF-β promotes heterogeneity and drug resistance in squamous cell carcinoma. Cell 160, 963–976 (2015).
Huggins, C. & Hodges, C. V. Studies on prostatic cancer. I. The effect of castration, of estrogen and androgen injection on serum phosphatases in metastatic carcinoma of the prostate. CA Cancer J. Clin. 22, 232–240 (1972).
Knudsen, K. E. & Scher, H. I. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin. Cancer Res. 15, 4792–4798 (2009).
Watson, P. A., Arora, V. K. & Sawyers, C. L. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat. Rev. Cancer 15, 701–711 (2015).
Arora, V. K. et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 155, 1309–1322 (2013).
Isikbay, M. et al. Glucocorticoid receptor activity contributes to resistance to androgen-targeted therapy in prostate cancer. Horm. Cancer 5, 72–89 (2014).
Beltran, H. et al. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 1, 487–495 (2011).
Beltran, H. et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016).
Zou, M. et al. Transdifferentiation as a mechanism of treatment resistance in a mouse model of castration-resistant prostate cancer. Cancer Discov. 7, 736–749 (2017).
Guo, C. C. et al. TMPRSS2-ERG gene fusion in small cell carcinoma of the prostate. Hum. Pathol. 42, 11–17 (2011).
Wang, H. T. et al. Neuroendocrine prostate cancer (NEPC) progressing from conventional prostatic adenocarcinoma: factors associated with time to development of NEPC and survival from NEPC diagnosis-a systematic review and pooled analysis. J. Clin. Oncol. 32, 3383–3390 (2014).
Nouri, M. et al. Therapy-induced developmental reprogramming of prostate cancer cells and acquired therapy resistance. Oncotarget 8, 18949–18967 (2017).
Mu, P. et al. SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88 (2017).
Ku, S. Y. et al. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 355, 78–83 (2017). This study and reference 50 were the first to identify the role of RB1 and TP53 in regulating tumour cell plasticity in prostate adenocarcinoma.
Zakowski, M. F., Ladanyi, M., Kris, M. G. & Memorial Sloan-Kettering Cancer Center Lung Cancer OncoGenome Group. EGFR mutations in small-cell lung cancers in patients who have never smoked. N. Engl. J. Med. 355, 213–215 (2006). This paper describes the first clinical case of small-cell lung transformation in lung adenocarcinoma.
Ferrer, L. et al. A brief report of transformation from NSCLC to SCLC: molecular and therapeutic characteristics. J. Thorac. Oncol. 14, 130–134 (2019).
Lee, J. K. et al. Clonal history and genetic predictors of transformation into small-cell carcinomas from lung adenocarcinomas. J. Clin. Oncol. 35, 3065–3074 (2017). This article identifies inactivation of both TP53 and RB1 as a predictive marker of small cell transformation in lung adenocarcinoma.
Marcoux, N. et al. EGFR-mutant adenocarcinomas that transform to small-cell lung cancer and other neuroendocrine carcinomas: clinical outcomes. J. Clin. Oncol. 37, 278–285 (2019).
Niederst, M. J. et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat. Commun. 6, 6377 (2015). This study discusses the role of RB1 in neuroendocrine transdifferentiation of lung adenocarcinomas.
Takegawa, N. et al. Transformation of ALK rearrangement-positive adenocarcinoma to small-cell lung cancer in association with acquired resistance to alectinib. Ann. Oncol. 27, 953–955 (2016).
Balla, A., Khan, F., Hampel, K. J., Aisner, D. L. & Sidiropoulos, N. Small-cell transformation of ALK-rearranged non-small-cell adenocarcinoma of the lung. Cold Spring Harb. Mol. Case Stud. 4, a002394 (2018).
Hobeika, C. et al. ALK-rearranged adenocarcinoma transformed to small-cell lung cancer: a new entity with specific prognosis and treatment? Per. Med. 15, 111–115 (2018).
Caumont, C. et al. Neuroendocrine phenotype as an acquired resistance mechanism in ALK-rearranged lung adenocarcinoma. Lung Cancer 92, 15–18 (2016).
Ou, S. I. et al. Dual occurrence of ALK G1202R solvent front mutation and small cell lung cancer transformation as resistance mechanisms to second generation ALK inhibitors without prior exposure to crizotinib. Pitfall of solely relying on liquid re-biopsy? Lung Cancer 106, 110–114 (2017).
Adelstein, D. J., Tomashefski, J. F. Jr., Snow, N. J., Horrigan, T. P. & Hines, J. D. Mixed small cell and non-small cell lung cancer. Chest 89, 699–704 (1986).
Norkowski, E. et al. Small-cell carcinoma in the setting of pulmonary adenocarcinoma: new insights in the era of molecular pathology. J. Thorac. Oncol. 8, 1265–1271 (2013).
Greenberg, N. M. et al. Prostate cancer in a transgenic mouse. Proc. Natl Acad. Sci. USA 92, 3439–3443 (1995).
Masumori, N. et al. A probasin-large T antigen transgenic mouse line develops prostate adenocarcinoma and neuroendocrine carcinoma with metastatic potential. Cancer Res. 61, 2239–2249 (2001).
Zhou, Z. et al. Synergy of p53 and Rb deficiency in a conditional mouse model for metastatic prostate cancer. Cancer Res. 66, 7889–7898 (2006).
Niederst, M. J. & Engelman, J. A. Bypass mechanisms of resistance to receptor tyrosine kinase inhibition in lung cancer. Sci. Signal. 6, re6 (2013).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Rubin, A. I., Chen, E. H. & Ratner, D. Basal-cell carcinoma. N. Engl. J. Med. 353, 2262–2269 (2005).
Sekulic, A. et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N. Engl. J. Med. 366, 2171–2179 (2012).
Robarge, K. D. et al. GDC-0449 – a potent inhibitor of the Hedgehog pathway. Bioorg. Med Chem. Lett. 19, 5576–5581 (2009).
Basset-Seguin, N. et al. Vismodegib in patients with advanced basal cell carcinoma (STEVIE): a pre-planned interim analysis of an international, open-label trial. Lancet Oncol. 16, 729–736 (2015).
Jain, S., Song, R. & Xie, J. Sonidegib: mechanism of action, pharmacology, and clinical utility for advanced basal cell carcinomas. Onco Targets Ther. 10, 1645–1653 (2017).
Yauch, R. L. et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 326, 572–574 (2009).
Sharpe, H. J. et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell 27, 327–341 (2015).
Atwood, S. X. et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell 27, 342–353 (2015).
Zhao, X. et al. A transposon screen identifies loss of primary cilia as a mechanism of resistance to SMO inhibitors. Cancer Discov. 7, 1436–1449 (2017).
Whitson, R. J. et al. Noncanonical hedgehog pathway activation through SRF-MKL1 promotes drug resistance in basal cell carcinomas. Nat. Med. 24, 271–281 (2018).
Sofen, H. et al. A phase II, multicenter, open-label, 3-cohort trial evaluating the efficacy and safety of vismodegib in operable basal cell carcinoma. J. Am. Acad. Dermatol. 73, 99–105.e1 (2015).
Lim, X. & Nusse, R. Wnt signaling in skin development, homeostasis, and disease. Cold Spring Harb. Perspect. Biol. 5, a008029 (2013).
Winder, M. & Viros, A. Mechanisms of drug resistance in melanoma. Handb Exp. Pharmacol. 249, 91–108 (2018). This review provides an overview of the mechanisms of acquired drug resistance in melanoma.
Manzano, J. L. et al. Resistant mechanisms to BRAF inhibitors in melanoma. Ann. Transl. Med 4, 237 (2016).
Ahmed, F. & Haass, N. K. Microenvironment-driven dynamic heterogeneity and phenotypic plasticity as a mechanism of melanoma therapy resistance. Front. Oncol. 8, 173 (2018).
Arozarena, I. & Wellbrock, C. Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer 19, 377–391 (2019).
Hoek, K. S. et al. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 68, 650–656 (2008).
Ahn, A., Chatterjee, A. & Eccles, M. R. The slow cycling phenotype: a growing problem for treatment resistance in melanoma. Mol. Cancer Ther. 16, 1002–1009 (2017).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Long, J. E. et al. Therapeutic resistance and susceptibility is shaped by cooperative multi-compartment tumor adaptation. Cell Death Differ. https://doi.org/10.1038/s41418-019-0310-0 (2019).
Levy, C., Khaled, M. & Fisher, D. E. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 12, 406–414 (2006).
Carreira, S. et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes Dev. 20, 3426–3439 (2006).
Saez-Ayala, M. et al. Directed phenotype switching as an effective antimelanoma strategy. Cancer Cell 24, 105–119 (2013).
Johannessen, C. M. et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature 504, 138–142 (2013).
Smith, M. P. et al. Inhibiting drivers of non-mutational drug tolerance is a salvage strategy for targeted melanoma therapy. Cancer Cell 29, 270–284 (2016).
Haq, R. et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc. Natl Acad. Sci. USA 110, 4321–4326 (2013).
Van Allen, E. M. et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 4, 94–109 (2014).
Smith, M. P. et al. Effect of SMURF2 targeting on susceptibility to MEK inhibitors in melanoma. J. Natl Cancer Inst. 105, 33–46 (2013).
Shi, H. et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 4, 80–93 (2014).
Muller, J. et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 5, 5712 (2014).
Konieczkowski, D. J. et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 4, 816–827 (2014).
Tsoi, J. et al. Multi-stage differentiation defines melanoma subtypes with differential vulnerability to drug-induced iron-dependent oxidative stress. Cancer Cell 33, 890–904.e5 (2018).
Menon, D. R. et al. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene 34, 4545 (2015).
Balaban, N. Q., Gerdes, K., Lewis, K. & McKinney, J. D. A problem of persistence: still more questions than answers? Nat. Rev. Microbiol. 11, 587–591 (2013).
Holden, D. W. Microbiology. Persisters unmasked. Science 347, 30–32 (2015).
Fisher, R. A., Gollan, B. & Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464 (2017).
Guler, G. D. et al. Repression of stress-induced line-1 expression protects cancer cell subpopulations from lethal drug exposure. Cancer Cell 32, 221–237.e13 (2017).
Touil, Y. et al. Colon cancer cells escape 5FU chemotherapy-induced cell death by entering stemness and quiescence associated with the c-Yes/YAP axis. Clin. Cancer Res. 20, 837–846 (2014).
Liau, B. B. et al. Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell 20, 233–246.e7 (2017).
Dardenne, E. et al. N-Myc induces an EZH2-mediated transcriptional program driving neuroendocrine prostate cancer. Cancer Cell 30, 563–577 (2016).
Kim, K. H. & Roberts, C. W. Targeting EZH2 in cancer. Nat. Med. 22, 128–134 (2016).
Varambally, S. et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624–629 (2002).
Clermont, P. L. et al. Polycomb-mediated silencing in neuroendocrine prostate cancer. Clin. Epigenetics 7, 40 (2015).
Murai, F. et al. EZH2 promotes progression of small cell lung cancer by suppressing the TGF-β-Smad-ASCL1 pathway. Cell Discov. 1, 15026 (2015).
Lapuk, A. V. et al. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J. Pathol. 227, 286–297 (2012).
Svensson, C. et al. REST mediates androgen receptor actions on gene repression and predicts early recurrence of prostate cancer. Nucleic Acids Res. 42, 999–1015 (2014).
Schoenherr, C. J. & Anderson, D. J. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267, 1360–1363 (1995).
Ballas, N., Grunseich, C., Lu, D. D., Speh, J. C. & Mandel, G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 (2005).
Lim, J. S. et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 545, 360–364 (2017).
Sarkar, A. & Hochedlinger, K. The Sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12, 15–30 (2013).
Buenrostro, J. D., Wu, B., Chang, H. Y. & Greenleaf, W. J. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–9 (2015).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Ferone, G. et al. SOX2 is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell 30, 519–532 (2016).
Bishop, J. L. et al. The master neural transcription factor BRN2 is an androgen receptor-suppressed driver of neuroendocrine differentiation in prostate cancer. Cancer Discov. 7, 54–71 (2017).
Sun, C. et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 508, 118–122 (2014).
Dravis, C. et al. Epigenetic and transcriptomic profiling of mammary gland development and tumor models disclose regulators of cell state plasticity. Cancer Cell 34, 466–482.e6 (2018).
Rajan, P. et al. Next-generation sequencing of advanced prostate cancer treated with androgen-deprivation therapy. Eur. Urol. 66, 32–39 (2014).
Li, X. et al. Prostate tumor progression is mediated by a paracrine TGF-β/Wnt3a signaling axis. Oncogene 27, 7118–7130 (2008).
Sun, Y. et al. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat. Med. 18, 1359–1368 (2012).
Bishop, J. L., Thaper, D. & Zoubeidi, A. The multifaceted roles of STAT3 signaling in the progression of prostate cancer. Cancers 6, 829–859 (2014).
Uysal-Onganer, P. et al. Wnt-11 promotes neuroendocrine-like differentiation, survival and migration of prostate cancer cells. Mol. Cancer 9, 55 (2010).
Chang, P. C. et al. Autophagy pathway is required for IL-6 induced neuroendocrine differentiation and chemoresistance of prostate cancer LNCaP cells. PLOS ONE 9, e88556 (2014).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437 (2013).
Zheng, H. et al. Therapeutic antibody targeting tumor- and osteoblastic niche-derived jagged1 sensitizes bone metastasis to chemotherapy. Cancer Cell 32, 731–747.e6 (2017).
Sharma, S. et al. Secreted protein acidic and rich in cysteine (sparc) mediates metastatic dormancy of prostate cancer in bone. J. Biol. Chem. 291, 19351–19363 (2016).
Keeratichamroen, S., Lirdprapamongkol, K. & Svasti, J. Mechanism of ECM-induced dormancy and chemoresistance in a549 human lung carcinoma cells. Oncol. Rep. 39, 1765–1774 (2018).
Hirata, E. et al. Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin β1/FAK signaling. Cancer Cell 27, 574–588 (2015).
Straussman, R. et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 487, 500–504 (2012).
Wilson, T. R. et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 487, 505–509 (2012).
Kaur, A. et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature 532, 250–254 (2016).
Roswall, P. et al. Microenvironmental control of breast cancer subtype elicited through paracrine platelet-derived growth factor-CC signaling. Nat. Med. 24, 463–473 (2018).
Ruffell, B. & Coussens, L. M. Macrophages and therapeutic resistance in cancer. Cancer Cell 27, 462–472 (2015).
Lee, G. T. et al. Macrophages induce neuroendocrine differentiation of prostate cancer cells via BMP6-IL6 Loop. Prostate 71, 1525–1537 (2011).
Smith, M. P. et al. The immune microenvironment confers resistance to MAPK pathway inhibitors through macrophage-derived TNFα. Cancer Discov. 4, 1214–1229 (2014).
Yao, Z. et al. TGF-β IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc. Natl Acad. Sci. USA 107, 15535–15540 (2010).
Qin, Y. et al. Hypoxia-driven mechanism of vemurafenib resistance in melanoma. Mol. Cancer Ther. 15, 2442–2454 (2016).
Liu, S., Kumar, S. M., Martin, J. S., Yang, R. & Xu, X. Snail1 mediates hypoxia-induced melanoma progression. Am. J. Pathol. 179, 3020–3031 (2011).
Das Thakur, M. et al. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 494, 251–255 (2013).
Tan, C. S., Gilligan, D. & Pacey, S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 16, e447–e459 (2015).
Masui, K. et al. A tale of two approaches: complementary mechanisms of cytotoxic and targeted therapy resistance may inform next-generation cancer treatments. Carcinogenesis 34, 725–738 (2013).
Robert, L., Ribas, A. & Hu-Lieskovan, S. Combining targeted therapy with immunotherapy. Can 1+1 equal more than 2? Semin. Immunol. 28, 73–80 (2016).
Terai, H. et al. ER stress signaling promotes the survival of cancer "persister cells" tolerant to EGFR tyrosine kinase inhibitors. Cancer Res. 78, 1044–1057 (2018).
Song, C. et al. Recurrent tumor cell-intrinsic and -extrinsic alterations during MAPKi-induced melanoma regression and early adaptation. Cancer Discov. 7, 1248–1265 (2017).
Kruidenier, L. et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488, 404–408 (2012).
Heinemann, B. et al. Inhibition of demethylases by GSK-J1/J4. Nature 514, E1–E2 (2014).
Kruidenier, L. et al. Kruidenier et al. reply. Nature 514, E2 (2014).
Johansson, C. et al. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat. Chem. Biol. 12, 539–545 (2016).
Yang, G. J. et al. Selective inhibition of lysine-specific demethylase 5A (KDM5A) using a rhodium(III) complex for triple-negative breast cancer therapy. Angew. Chem. Int. Ed. 57, 13091–13095 (2018).
Yang, G. J., Ko, C. N., Zhong, H. J., Leung, C. H. & Ma, D. L. Structure-based discovery of a selective KDM5A inhibitor that exhibits anti-cancer activity via inducing cell cycle arrest and senescence in breast cancer cell lines. Cancers 11, E92 (2019).
Bradner, J. E., Hnisz, D. & Young, R. A. Transcriptional addiction in cancer. Cell 168, 629–643 (2017).
Kwiatkowski, N. et al. Targeting transcription regulation in cancer with a covalent CDK7 inhibitor. Nature 511, 616–620 (2014).
Rusan, M. et al. Suppression of adaptive responses to targeted cancer therapy by transcriptional repression. Cancer Discov. 8, 59–73 (2018).
Zawistowski, J. S. et al. Enhancer remodeling during adaptive bypass to MEK inhibition is attenuated by pharmacologic targeting of the P-TEFb complex. Cancer Discov. 7, 302–321 (2017).
Knoechel, B. et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat. Genet. 46, 364–370 (2014).
Smith, P. C. & Keller, E. T. Anti-interleukin-6 monoclonal antibody induces regression of human prostate cancer xenografts in nude mice. Prostate 48, 47–53 (2001).
Wallner, L. et al. Inhibition of interleukin-6 with CNTO328, an anti-interleukin-6 monoclonal antibody, inhibits conversion of androgen-dependent prostate cancer to an androgen-independent phenotype in orchiectomized mice. Cancer Res. 66, 3087–3095 (2006).
Dorff, T. B. et al. Clinical and correlative results of SWOG S0354: a phase II trial of CNTO328 (siltuximab), a monoclonal antibody against interleukin-6, in chemotherapy-pretreated patients with castration-resistant prostate cancer. Clin. Cancer Res. 16, 3028–3034 (2010).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT00433446 (2013).
Liu, J. et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl Acad. Sci. USA 110, 20224–20229 (2013).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT01351103 (2011).
Kahn, M. Can we safely target the WNT pathway? Nat. Rev. Drug Discov. 13, 513–532 (2014).
Eberl, M. et al. Tumor architecture and notch signaling modulate drug response in basal cell carcinoma. Cancer Cell 33, 229–243.e4 (2018).
Kim, I. S. et al. Microenvironment-derived factors driving metastatic plasticity in melanoma. Nat. Commun. 8, 14343 (2017).
Smith, M. P. et al. Targeting endothelin receptor signalling overcomes heterogeneity driven therapy failure. EMBO Mol. Med. 9, 1011–1029 (2017).
Hughes, V. S. & Siemann, D. W. Have clinical trials properly assessed c-Met inhibitors? Trends Cancer 4, 94–97 (2018).
Graff, J. N. et al. Early evidence of anti-PD-1 activity in enzalutamide-resistant prostate cancer. Oncotarget 7, 52810–52817 (2016).
Wilson, C. et al. AXL inhibition sensitizes mesenchymal cancer cells to antimitotic drugs. Cancer Res. 74, 5878–5890 (2014).
Sheridan, C. First Axl inhibitor enters clinical trials. Nat. Biotechnol. 31, 775–776 (2013).
BerGenBio. BerGenBio reports promising BGB324 phase I/II monotherapy data in patients with lung cancer at the EORTC-NCI-AACR molecular targets and cancer therapeutics symposium. BerGenBio https://www.bergenbio.com/bergenbio-reports-promising-bgb324-phase-iii-monotherapy-data-in-patients-with-lung-cancer-at-the-eortc-nci-aacr-molecular-targets-and-cancer-therapeutics-symposium/ (2016).
Boshuizen, J. et al. Cooperative targeting of melanoma heterogeneity with an AXL antibody-drug conjugate and BRAF/MEK inhibitors. Nat. Med. 24, 203–212 (2018).
Viswanathan, V. S. et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 (2017).
Hangauer, M. J. et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250 (2017).
Friedmann Angeli, J. P. et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191 (2014).
Yoo, S. E. et al. Gpx4 ablation in adult mice results in a lethal phenotype accompanied by neuronal loss in brain. Free. Radic Biol. Med. 52, 1820–1827 (2012).
Eaton, J. K. et al. Targeting a therapy-resistant cancer cell state using masked electrophiles as GPX4 inhibitors. Preprint at bioRxiv https://doi.org/10.1101/376764 (2018).
Knutson, S. K. et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 13, 842–854 (2014).
Vaswani, R. G. et al. Identification of (R)-N-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J. Med. Chem. 59, 9928–9941 (2016).
Constellation Pharmaceuticals. Constellation Pharmaceuticals announces first patient dosed in phase 1b/2 PROSTAR combination study of CPI-1205 in advanced form of prostate cancer. Constell. Pharm. https://www.constellationpharma.com/constellation-pharmaceuticals-announces-first-patient-dosed-phase-1b-2-prostar-combination-study-cpi-1205-advanced-form-prostate-cancer/ (2017).
US National Library of Medicine. ClinicalTrials.gov https://www.clinicaltrials.gov/ct2/show/NCT02082977 (2017).
Gu, S., Cui, D., Chen, X., Xiong, X. & Zhao, Y. PROTACs: an emerging targeting technique for protein degradation in drug discovery. Bioessays 40, e1700247 (2018).
Sievers, Q. L. et al. Defining the human C2H2 zinc finger degrome targeted by thalidomide analogs through CRBN. Science 362, eaat0572 (2018).
Massague, J. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13, 616–630 (2012).
Akhurst, R. J. & Hata, A. Targeting the TGFβ signalling pathway in disease. Nat. Rev. Drug Discov. 11, 790–811 (2012).
Pattabiraman, D. R. et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 351, aad3680 (2016).
Nieto, M. A., Huang, R. Y., Jackson, R. A. & Thiery, J. P. EMT: 2016. Cell 166, 21–45 (2016).
Ocana, O. H. et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 22, 709–724 (2012).
Tsai, J. H., Donaher, J. L., Murphy, D. A., Chau, S. & Yang, J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22, 725–736 (2012).
Ishay-Ronen, D. et al. Gain fat-lose metastasis: converting invasive breast cancer cells into adipocytes inhibits cancer metastasis. Cancer Cell 35, 17–32.e6 (2019).
Waddington, C. H. The Strategy of the Genes; a Discussion of Some Aspects of Theoretical Biology. (Allen & Unwin, 1957).
Rajagopal, J. & Stanger, B. Z. Plasticity in the adult: how should the Waddington diagram be applied to regenerating tissues? Dev. Cell 36, 133–137 (2016).
Blanpain, C. & Fuchs, E. Stem cell plasticity. Plasticity of epithelial stem cells in tissue regeneration. Science 344, 1242281 (2014).
Paksa, A. & Rajagopal, J. The epigenetic basis of cellular plasticity. Curr. Opin. Cell Biol. 49, 116–122 (2017).
Ito, M. et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11, 1351–1354 (2005).
Tetteh, P. W. et al. Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell 18, 203–213 (2016).
van Es, J. H. et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nat. Cell Biol. 14, 1099–1104 (2012).
de Sousa e Melo, F. & de Sauvage, F. J. Cellular plasticity in intestinal homeostasis and disease. Cell Stem Cell 24, 54–64 (2019).
Barrett, N. R. The lower esophagus lined by columnar epithelium. Surgery 41, 881–894 (1957).
Van Keymeulen, A. et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189–193 (2011).
Battula, V. L. et al. Epithelial-mesenchymal transition-derived cells exhibit multilineage differentiation potential similar to mesenchymal stem cells. Stem Cells 28, 1435–1445 (2010).
Krebs, A. M. et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 19, 518–529 (2017).
McKeithen, D., Graham, T., Chung, L. W. & Odero-Marah, V. Snail transcription factor regulates neuroendocrine differentiation in LNCaP prostate cancer cells. Prostate 70, 982–992 (2010).
Akamatsu, S. et al. The placental gene PEG10 promotes progression of neuroendocrine prostate cancer. Cell Rep. 12, 922–936 (2015).
Park, K. S. et al. Characterization of the cell of origin for small cell lung cancer. Cell Cycle 10, 2806–2815 (2011).
Sutherland, K. D. et al. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 19, 754–764 (2011).
Sutherland, K. D. et al. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc. Natl Acad. Sci. USA 111, 4952–4957 (2014).
Mainardi, S. et al. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc. Natl Acad. Sci. USA 111, 255–260 (2014).
Miettinen, P. J. et al. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature 376, 337–341 (1995).
Youssef, K. K. et al. Adult interfollicular tumour-initiating cells are reprogrammed into an embryonic hair follicle progenitor-like fate during basal cell carcinoma initiation. Nat. Cell Biol. 14, 1282–1294 (2012).
Choi, N., Zhang, B., Zhang, L., Ittmann, M. & Xin, L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell 21, 253–265 (2012).
Xin, L. Cells of origin for cancer: an updated view from prostate cancer. Oncogene 32, 3655–3663 (2013).
Suraweera, A., O'Byrne, K. J. & Richard, D. J. Combination therapy with histone deacetylase inhibitors (HDACi) for the treatment of cancer: achieving the full therapeutic potential of HDACi. Front. Oncol. 8, 92 (2018).
Han, J. Y. et al. Phase I/II study of gefitinib (Iressa((R))) and vorinostat (IVORI) in previously treated patients with advanced non-small cell lung cancer. Cancer Chemother. Pharmacol. 75, 475–483 (2015).
Reguart, N. et al. Phase I/II trial of vorinostat (SAHA) and erlotinib for non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) mutations after erlotinib progression. Lung Cancer 84, 161–167 (2014).
Wang, L. et al. An acquired vulnerability of drug-resistant melanoma with therapeutic potential. Cell 173, 1413–1425.e14 (2018).
Ferrari, A. C. et al. Epigenetic therapy with panobinostat combined with bicalutamide rechallenge in castration-resistant prostate cancer. Clin. Cancer Res. 25, 52–63 (2019).
Banerji, U. et al. A phase I pharmacokinetic and pharmacodynamic study of CHR-3996, an oral class I selective histone deacetylase inhibitor in refractory solid tumors. Clin. Cancer Res. 18, 2687–2694 (2012).
Lin, H. et al. Small molecule KDM4s inhibitors as anti-cancer agents. J. Enzyme Inhib. Med. Chem. 33, 777–793 (2018).
Tumber, A. et al. Potent and selective KDM5 inhibitor stops cellular demethylation of H3K4me3 at transcription start sites and proliferation of MM1S myeloma cells. Cell Chem. Biol. 24, 371–380 (2017).
Gale, M. et al. Screen-identified selective inhibitor of lysine demethylase 5A blocks cancer cell growth and drug resistance. Oncotarget 7, 39931–39944 (2016).
Liang, J. et al. From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors. Bioorg. Med. Chem. Lett. 27, 2974–2981 (2017).
Patel, H. et al. ICEC0942, an orally bioavailable selective inhibitor of CDK7 for cancer treatment. Mol. Cancer Ther. 17, 1156–1166 (2018).
Syros Pharmaceuticals. Syros presents new preclinical PK and PD data for SY-1365, its first-in-class selective CDK7 inhibitor, at AACR-NCI-EORTC conference. Syros Pharma. https://ir.syros.com/press-releases/detail/106/syros-presents-new-preclinical-pk-and-pd-data-for-sy-1365 (2017).
Gerlach, D. et al. The novel BET bromodomain inhibitor BI 894999 represses super-enhancer-associated transcription and synergizes with CDK9 inhibition in AML. Oncogene 37, 2687–2701 (2018).
Pervaiz, M., Mishra, P. & Gunther, S. Bromodomain drug discovery - the past, the present, and the future. Chem. Rec. 18, 1808–1817 (2018).
Hogg, S. J. et al. BET-bromodomain inhibitors engage the host immune system and regulate expression of the immune checkpoint ligand PD-L1. Cell Rep. 18, 2162–2174 (2017).
Hudes, G. et al. A phase 1 study of a chimeric monoclonal antibody against interleukin-6, siltuximab, combined with docetaxel in patients with metastatic castration-resistant prostate cancer. Invest. New Drugs 31, 669–676 (2013).
Jiang, J. et al. A novel porcupine inhibitor blocks WNT pathways and attenuates cardiac hypertrophy. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3459–3467 (2018).
Proffitt, K. D. et al. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res. 73, 502–507 (2013).
Jackson, H. et al. Novel bispecific domain antibody to LRP6 inhibits Wnt and R-spondin ligand-induced wnt signaling and tumor growth. Mol Cancer Res 14, 859–868 (2016).
Gong, Y. et al. Wnt isoform-specific interactions with coreceptor specify inhibition or potentiation of signaling by LRP6 antibodies. PLOS ONE 5, e12682 (2010).
Tamagnone, L., Zacchigna, S. & Rehman, M. Taming the Notch transcriptional regulator for cancer therapy. Molecules 23, E431 (2018).
Krop, I. et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J. Clin. Oncol. 30, 2307–2313 (2012).
Messersmith, W. A. et al. A phase I, dose-finding study in patients with advanced solid malignancies of the oral γ-secretase inhibitor PF-03084014. Clin. Cancer Res. 21, 60–67 (2015).
Italiano, A. et al. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumours: a first-in-human, open-label, phase 1 study. Lancet Oncol. 19, 649–659 (2018).
Kung, P. P. et al. Optimization of orally bioavailable enhancer of zeste homolog 2 (EZH2) inhibitors using ligand and property-based design strategies: identification of development candidate (R)-5,8-dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2- dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497). J. Med. Chem 61, 650–665 (2018).
Acknowledgements
The authors thank Felipe de Sousa e Melo, Ciara Metcalfe, Xin Ye and Bob Yauch for their valuable comments and suggestions on the manuscript. We also thank Allison Bruce for drawing the figures.
Author information
Authors and Affiliations
Contributions
S.B. and F.J.d.S. researched data for the article, provided substantial contribution to discussions of the content, wrote the article, and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors are employees of Genentech and own shares of Roche.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Complete response
-
Reduction in tumour burden measured by radiological imaging or biopsy typically corresponding to at least 99% decrease.
- Minimal residual disease
-
(MRD). Tumour cells that remain once a patient has achieved complete response as measured through radiological imaging or biopsy.
- Apical–basal polarity
-
Polarity typical of epithelial cells, in which the apical surface faces the lumen and the basal side faces the basement membrane.
- Transdetermination
-
The process of lineage conversion, in which a tissue-specific progenitor cell population transforms into the progenitor cell population of a distinct tissue.
- Cell lineage
-
Developmental history of a differentiated cell tracing its cellular origins.
- Transdifferentiation
-
Direct conversion of one differentiated cell type into another without going through a progenitor state.
- Ferroptosis
-
Type of programmed cell death that is dependent on iron and characterized by the accumulation of lipid peroxide chemical species.
- Dedifferentiation
-
The process of lineage reversion in which differentiated cells acquire features of more immature cells within the same lineage.
Rights and permissions
About this article
Cite this article
Boumahdi, S., de Sauvage, F.J. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat Rev Drug Discov 19, 39–56 (2020). https://doi.org/10.1038/s41573-019-0044-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41573-019-0044-1
This article is cited by
-
Ribosome profiling: a powerful tool in oncological research
Biomarker Research (2024)
-
Unveiling the mechanisms and challenges of cancer drug resistance
Cell Communication and Signaling (2024)
-
Zeb1-controlled metabolic plasticity enables remodeling of chromatin accessibility in the development of neuroendocrine prostate cancer
Cell Death & Differentiation (2024)
-
Targeting initial tumour–osteoclast spatiotemporal interaction to prevent bone metastasis
Nature Nanotechnology (2024)
-
HMGA2 alleviates ferroptosis by promoting GPX4 expression in pancreatic cancer cells
Cell Death & Disease (2024)