Abstract
Autophagy may control the de novo refractoriness of HER2 gene-amplified breast carcinomas to the monoclonal antibody trastuzumab (Herceptin). Tumor cells originally obtained from a patient who rapidly progressed on trastuzumab ab initio display increased cellular levels of the LC3-II protein—a finding that correlates with increased numbers of autophagosomes—and decreased levels of the autophagy receptor p62/SQSTM1, a protein selectively degraded by autophagy. Trastuzumab-refractory cells are in a state of “autophagy addiction” because genetic ablation of autophagy-specific genes (ATG8, ATG5, ATG12) notably reduces intrinsic refractoriness to trastuzumab. When the anti-malarial lysosomotropic drug chloroquine impedes autophagic resolution of the accumulation of autophagolysosomes formed in the presence of trastuzumab, cells commit to die by apoptosis. Accordingly, combination treatment with trastuzumab and chloroquine radically suppresses tumor growth by > 90% in a tumor xenograft completely refractory to trastuzumab. Adding chloroquine to trastuzumab-based regimens may therefore improve outcomes among women with autophagy-addicted HER2-positive breast cancer.
Similar content being viewed by others
Introduction
“The most fruitful basis for the discovery of a new drug is to start with an old drug”1. This statement from the renowned pharmacologist and Nobel laureate James Black properly defines the repurposing or repositioning approach of developing a known drug for another clinical purpose2. The repurposing approach may overcome the enormous problems involved in producing new anti-cancer drugs following the traditional approach of de novo drug discovery and development; this process can take an average of 15 years and several hundred million dollars to move from an idea to a marketed drug2,3. Scanning the existing pharmacopoeia for repositioning candidates can be a very effective way to develop new oncology therapeutics, as the pharmacokinetics and safety profiles of many existing drugs have been studied and these drugs often have already been approved for human use by regulatory agencies (FDA, MEA and MHLW). In this scenario, any “old drug” can be rapidly evaluated for “new uses” in phase II cancer clinical trials.
One of the well-known repositioning success stories relates to the (re)use of chloroquine, a well-known 5-aminoquinoline drug that is widely used for the prophylactic treatment of malaria4, as part of a combinational therapy for cancer. After six decades of use, chloroquine remains the drug of choice for malaria chemotherapy because it is effective, it has low toxicity in humans and it is inexpensive5. In its unprotonated form, chloroquine can diffuse across cell membranes to become protonated and accumulate in acidic organelles such as lysosomes6. This lysosomotropic property has been recently used to redefine chloroquine and its derivatives as late-phase inhibitors of macroautophagy (herein referred to as autophagy), an evolutionarily conserved cellular process by which cells sequester a portion of the cytoplasm and organelles into double-membraned vesicles that subsequently fuse with lysosomes for degradation of the enclosed materials7,8,9,10. Autophagy is recognized as a crucial cell survival pathway that enables tumor cells to overcome stressors in the tumor microenvironment as well as injuries caused by treatments such as endocrine therapy, chemotherapy and radiation therapy11,12,13,14,15. Because the abrogation of autophagy via knockdown of autophagy-related molecules potentiates the re-sensitization of therapy-resistant cancer cells to conventional cancer therapies, there has been great interest in developing clinically relevant autophagy inhibitors.
Chloroquine's ability to block autophagy by inhibiting lysosomal proteases and preventing autophagosome-lysosome fusion events has established chloroquine as the most widely used drug to inhibit autophagy in vitro and in vivo14,15,16,17,18,19,20. Indeed, chloroquine and its derivatives are currently the only inhibitors used for treatment of cancer patients and more than 20 clinical trials using chloroquine or hydroxychloroquine are now testing whether the pharmacological inhibition of autophagy in a clinical setting can increase the effectiveness of existing cancer therapies (http://clinicaltrials.gov/ct2/results?term=autophagy+and+cancer&Search=&Search=Search)21,22. All human clinical trials exploring autophagy inhibition as a therapeutic strategy have used chloroquine or its derivative hydroxychloroquine due to its long track record of safety in human patients; however, whether chloroquine and its derivatives represent the most efficacious drugs for inhibiting autophagy remains highly debatable. First, the high doses of chloroquine required to achieve tumor inhibition in humans might not be ideal due to the pharmacology of the drug. Accordingly, the combination of the chloroquine derivative hydroxychloroquine with chemotherapy, proteasome inhibitors, mTOR inhibitors, and/or radiation has been shown to result in low response rates in preliminary clinical trials22, indicating that hydroxychloroquine is not a potent autophagy inhibitor at clinically tolerable doses. Moreover, it has been recently demonstrated that chloroquine-mediated chemosensitization to therapy appears to be an autophagy-independent event (i.e., chloroquine was able to sensitize cancer cells to treatment even when autophagy was blocked upstream of autophagosome formation)21. Together, these findings strongly demonstrate a need to better understand the specific molecular settings in which autophagy is or is not cytoprotective and to develop more selective autophagy inhibitors (e.g., Lys05 and Spautin-1)23,24,25 to maximize the benefits of pharmacologically targeting autophagy during cancer therapy21.
Despite the above-mentioned unfavorable findings, we have recently envisioned that chloroquine-based, “customized” combinatorial approaches could prove to be more effective and autophagy-specific when combined with molecularly targeted drugs in biomarker-identified patient subpopulations whose carcinomas are addicted to specific, autophagy-related mechanisms of malignant transformation. Specifically, our objective was to overcome or circumvent the highly prevalent inherent (i.e., primary) resistance of HER2-overexpressing breast carcinomas to the anti-HER2 monoclonal antibody trastuzumab (Herceptin). We utilized the JIMT-1 cell line, a low-passage pleural effusion explant originally obtained from a breast cancer patient who rapidly progressed on trastuzumab ab initio. This cell line constitutes a unique patient-derived model system for studying the intrinsic26,27,28,29,30,31,32,33,34,35,36 and immune-mediated37,38 growth-inhibitory mechanisms of trastuzumab in vitro and in xenograft models. We first assessed whether constitutive activation of protective autophagy in HER2 gene-amplified breast carcinomas could operate as a novel mechanism causing de novo (primary) refractoriness to trastuzumab. Given that trastuzumab's mechanism of action involves HER2 internalization and recycling via chloroquine-targeted lysosomal pathway-dependent degradation, we then evaluated whether adding chloroquine to trastuzumab might uncover a previously unrecognized synthetic lethal interaction. Here, we present the first experimental evidence showing that protective autophagy regulates primary resistance to trastuzumab and that adding chloroquine to trastuzumab-based regimens may significantly improve outcomes among women with autophagy-addicted, HER2-positive breast cancer.
Results
Trastuzumab-refractory JIMT-1 breast cancer cells exhibit constitutively enhanced autophagic vesicle content and an increased autophagic flux
A well-accepted hallmark of autophagy induction is the lipidation of the microtubule-associated protein 1 light chain 3 (LC3), the mammalian homolog of Atg8 termed LC3-II, which becomes membrane associated and actively participates in autophagosome formation. Because LC3-II levels (normalized to actin or tubulin loading controls) correlate well with autophagosome numbers (or more precisely, the amount of autophagic membrane labeled with LC3-II)39,40,41, we first compared the baseline conversion status of endogenous LC3 to LC3-II in trastuzumab-refractory JIMT-1 cells to that in trastuzumab-sensitive SKBR3 cells. Immunoblotting using an antibody against LC3 clearly revealed that trastuzumab-refractory JIMT-1 cells constitutively contained greater amounts of the hydrophobic form of LC3 (LC3-II) relative to trastuzumab-sensitive SKBR3 cells ( Figure 1A , left panels). Based on the assumption that the lysosomal degradation capacity remains constant in both cell lines, the cellular accumulation of LC3-II can be directly interpreted as a consequence of autophagy activation (i.e., increased autophagosome formation) in trastuzumab-refractory versus trastuzumab-sensitive cells. However, an increase in the cellular content of LC3-II can also be due to a block in autophagosome-lysosome fusion or lysosomal degradation, which results in the impairment of autophagosome maturation. To correctly interpret the LC3-II immunoblot data, we pharmacologically clamped the LC3-II autophagosome degradation using saturating concentrations of the lysosomal proton inhibitors bafilomycin A1 and chloroquine42. Given that treatment with either bafilomycin A1 or chloroquine blocks lysosomal degradation of autophagosomes, this approach allows an estimation of the total amount of “converted” LC3-II during autophagy blockade. We found that bafilomycin A1- and chloroquine-treated cells showed an increase in the total amount of LC3-II, strongly suggesting that a bona fide increase in autophagic flux occurs in trastuzumab-refractory JIMT1 cells when compared to trastuzumab-sensitive SKBR3 cells ( Figure 1A , left panels). The fact that quantitative real-time PCR (qRT-PCR)-based analyses of LC3 mRNA expression failed to detect significant upregulation43 definitely suggested that trastuzumab-refractory JIMT-1 cells, but not trastuzumab-sensitive SKBR3 cells, naturally exhibit constitutive enhancement of the net incorporation of LC3-II into autophagosomal membranes.
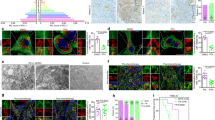
Autophagic flux dynamics in trastuzumab-sensitive and -refractory breast cancer cells.
(A). Left (immunoblotting). Autophagosome formation in whole cell lysates of trastuzumab-sensitive SKBR3 cells and trastuzumab-refractory JIMT-1 cells was assessed with Western blot analyses using a LC3 antibody. Top band (18 kDa) represents LC3-I. Bottom band (16 kDa) represents LC3-II, a typical marker of autophagosomes. Autophagic degradation in whole cell lysates of trastuzumab-sensitive SKBR3 cells and trastuzumab-refractory JIMT-1 cells was detected with Western blot analyses using a p62/SQSTM1 antibody. Immunoblotting band (64 kDa) represents p62/SQSTM1, a selective substrate of autophagy. Conversion of cytosolic LC3-I to autophagosome-associated LC3-II was assessed in the presence of chloroquine that, as a weak base, raises the lysosomal pH and thereby, like bafilomycin A1, inhibits late steps of autophagy (i.e., degradation of the cargo in autophagolysosomes). Figures show representative immunoblotting analyses of LC3 and p62/SQSTM1. Equivalent results were obtained in three independent experiments. Right (immunofluorescence). Images show representative portions of SKBR3 and JIMT-1 cell cultures that were captured using different channels for LC3 (red), p62 (green) and Hoechst 33258 (blue) with a 20 × objective and merged on a BD Pathway™ 855 Bioimager System using BD Attovision™ software. Scale bar = 25 μm. (B). Images show representative untreated and trastuzumab-treated whole populations of SKBR3 and JIMT-1 cells growing in individual wells that were captured using different channels for LC3 (red), p62/SQSTM1 (green) and Hoechst 33258 (blue) as a 4 × 4 montage with 20 × objective on a BD Pathway™ 855 Bioimager System and merged using BD Attovision™ software. Scale bar = 200 μm. White arrows indicate strong colocalization of LC3 and p62/SQSTM1 in subgroups of trastuzumab-treated JIMT-1 cells.
Another well-recognized hallmark of autophagy is the redistribution of LC3 from a diffuse cytoplasmic localization to a characteristic punctate cytoplasmic pattern that reflects the recruitment of LC3-II to autophagic vesicles. When LC3-II-vesicle formation was monitored by indirect immunofluorescence using an automated confocal imaging approach, we concluded that subcellular redistribution of endogenous LC3 into discrete cytoplasmic dots was markedly increased in individual JIMT-1 cells compared with individual SKBR3 cells ( Figure 1A , right panels). Whereas trastuzumab-sensitive SKBR3 cells exhibited a homogeneous but weak cytoplasmic staining of LC3, which is consistent with the basal distribution of LC3-I and typical of low-level or no autophagosome formation, LC3 localization notably changed from diffuse to a punctate or dotted pattern in trastuzumab-refractory JIMT-1 cells. High-content imaging of whole JIMT-1 cell populations growing as individual wells (captured as 4 × 4 montages) clearly revealed that > 90% of trastuzumab-refractory JIMT-1 cells exhibited intense punctate LC3 fluorescence, whereas a significantly lower percentage of trastuzumab-sensitive SKBR3 cells exhibited LC3-containing vesicles ( Figure 1B , right panels). Indeed, the dynamics of autophagosome formation in JIMT-1 cells, which exhibit de novo (primary) resistance to trastuzumab, was notably comparable to that observed in pre-clinical models of acquired (secondary) resistance to trastuzumab obtained upon continuous exposure of SKBR3 cells to clinically relevant doses of trastuzumab for more than 10 months ( Supplementary Figure 1 )44.
To determine more directly the degree of activated autophagy in JIMT-1 cells, we assessed the integration of LC3 into autophagosomes using a GFP-LC3 reporter, a standard assay to measure active autophagy. GFP-LC3 dots were rare in cultures of trastuzumab-sensitive SKBR3 cells ( Supplementary Figure 2 ), with evidence of GFP-LC3 puncta in less than 5% of these cells, consistent with the findings of others that high levels of autophagy are typically present in vitro only when cultured cells are deprived of essential nutrients or placed under other stresses. However, GFP-LC3 puncta were detected in 50–70% of trastuzumab-refractory JIMT-1 cells, indicating a high degree of autophagy activation ( Supplementary Figure 2 ). JIMT-1 cells grown in nutrient-rich conditions also had significantly elevated numbers of puncta per cell as compared with SKBR3 cells. When SKBR3 and JIMT-1 cells were cultured under normal growth conditions and the presence of LC3 dots was determined in the absence or presence of trastuzumab, treatment with trastuzumab further increased the number of GFP-LC3 puncta in JIMT-1 cells. Although the above-mentioned findings were consistent with a constitutive increase in flux through autophagic clearance pathways in trastuzumab-refractory JIMT-1 cells in a cell-autonomous manner, we adopted additional complementary experimental strategies to unambiguously distinguish whether increases in LC3 II-marked autophagosomes reflected either the induction of autophagy or the inhibition of autophagosome clearance. First, we performed preliminary indirect immunofluorescence by staining for the lysosome marker lysosomal-associated membrane protein-1 (LAMP-1) to determine whether the increase in autophagosome-related LC3 was a consequence of lysosomal dysfunction preventing the clearance of autophagosomes. We found that lysosome abundance in JIMT-1 cells was increased relative to that in SKBR3 cells (data not shown), suggesting that diminished lysosomal function did not contribute to autophagosome accumulation in trastuzumab-refractory JIMT-1 cells. Second, when high levels of the lipidated form of LC3 are associated with impairment in autophagosome maturation, this phenomenon is accompanied by a marked increase in the cellular content of the specific autophagy receptor molecule SQSTM1/p62, which can actively link autophagy targets with autophagosomal membranes45,46,47,48,49. The cellular content of SQSTM1/p62 negatively correlates with autophagic flux because SQSTM1/p62 is itself degraded by autophagy, as it serves to link ubiquitinated proteins to the autophagic machinery, thereby enabling their degradation in the lysosome. Therefore, a reduction in SQSTM1/p62 protein levels together with increased LC3-II levels characterizes the occurrence of a real increase in the autophagic flux. Immunoblotting studies in whole cell lysates confirmed that the baseline content of the SQSTM1/p62 protein was clearly decreased in trastuzumab-refractory JIMT-1 cells when compared with that in trastuzumab-sensitive SKBR3 cells ( Figure 1A , left panels). High content imaging of whole JIMT-1 cell populations growing in individual wells confirmed that most cells displayed a significant decrease in the amount of SQSTM1/p62 per individual cell when compared with SKBR3 cell populations ( Figure 1A , right panels; Figure 1B , left panels). Given that the relative mRNA levels of SQSTM1/p62 were not significantly altered as measured by qRT-PCR43, these findings indicate that the decreased cellular content of SQSTM1/p62 indirectly reflects enhanced autophagic degradation in trastuzumab-refractory JIMT-1 cells.
Autophagy addiction underlies primary resistance to trastuzumab
To determine whether the autophagy machinery of JIMT-1 cells performed a pro-survival function once the cells were challenged with the anti-HER2 monoclonal antibody trastuzumab, we monitored whether autophagy-specific (ATG) genes that are required for autophagosome formation functioned as indispensable controllers of de novo resistance to trastuzumab. We used lentivirus-mediated small hairpin (sh)RNA knockdown of the autophagy-related gene ATG8/LC3 (isoform LC3B). Together with ATG12, ATG8/LC3 functions in one of the ubiquitin-like conjugation systems required for autophagosome formation50,51,52,53. To control for any off-target effects of shRNA transduction, negative control lentiviral particles were used. Following stable transduction with negative control shRNA and ATG8/LC3 shRNA, immunoblotting of whole cell lysates confirmed that the steady-state expression of the ATG8/LC3 protein was efficiently knocked down54. We then assessed the impact of shRNA-driven genetic ablation of ATG8/LC3 on the growth inhibitory activity of trastuzumab. The ability of shRNA-mediated silencing of ATG8/LC3 to inhibit autophagy was demonstrated by the enhanced accumulation of p62/SQSTM1 and the reduced frequency of cells displaying LC3-positive autophagic vesicle formation ( Figure 2A , left panels). Importantly, whereas both parental JIMT1 cells and negative control shRNA-transduced JIMT-1 cells were completely refractory of trastuzumab, a small but significant increase in trastuzumab efficacy was observed in ATG8/LC3 shRNA-transduced JIMT-1 derivatives ( Figure 2A , right panels). The almost insignificant loss of cell viability induced by trastuzumab in both parental and negative control shRNA-transduced cells (i.e., 1% reduction at 1 μg/mL trastuzumab and 8% reduction at 100 μg/mL trastuzumab) were significantly augmented in ATG8/LC3 shRNA/JIMT1 cells (i.e., from 19% reduction at 1 μg/mL trastuzumab to 30% reduction at 100 μg/mL trastuzumab).
Genetic ablation of the autophagy machinery: Impact on primary resistance to HER targeting drugs.
(A). Immunofluorescence images. Functional validation of the ATG8/LC3 gene silencing in trastuzumab-refractory JIMT-1 cells. Images show representative portions of JIMT-1 parental (WT), control shRNA-JIMT-1 and ATG8/LC3 shRNA-JIMT-1 cell cultures that were captured for p62/SQSTM1 (green, left panels) or LC3 (green, right panels) and Hoechst 33258 (blue) with a 20 × objective and merged on a BD Pathway™ 855 Bioimager System using BD Attovision™ software. Scale bar = 25 μm. Graph. The metabolic status of JIMT-1 parental cells, control shRNA-JIMT-1 cells and ATG8/LC3 shRNA-JIMT-1 cells treated with graded concentrations trastuzumab was evaluated using MTT-based cell viability assays, followed by the generation of dose-response graphs depicting the% of untreated cells (untreated control cells = 100% cell viability). The results are presented as the means (columns) and 95% confidence intervals (bars) of three independent experiments performed in triplicate. (B). The metabolic status of JIMT-1 parental (WT), control shRNA-JIMT-1 and ATG8/LC3 shRNA-JIMT-1 cells treated with graded concentrations of gefitinib, erlotinib and lapatinib was evaluated using MTT-based cell viability assays, followed by the generation of dose-response graphs depicting the% of untreated cells (untreated control cells = 100% cell viability; figures not shown). Bars representing mean IC50 values in each cell line illustrate the degree of sensitivity to HER1/2-targeting drugs. Error bars show 95% confidence intervals of three independent experiments performed in triplicate.
HER2 gene-amplified JIMT-1 cells exhibit not only primary resistance to trastuzumab but also an intrinsic cross-resistance to multiple HER1/2 inhibitors34. Therefore, we speculated that shRNA-driven genetic ablation of autophagy addiction in JIMT-1 cells should result in a significantly increased efficacy of the small molecule HER1/2 tyrosine kinase inhibitors (TKIs) gefitinib, erlotinib and lapatinib ( Figure 2B ). The gefitinib's half maximal (50%) inhibitory concentration (IC) value decreased from 8.0 ± 1.5 μmol/L and 7.8 ± 1 μmol/L in parental and negative control shRNA/JIMT-1 cells, respectively, to 3.2 ± 0.5 μmol/L (p < 0.05) in ATG8/LC3 shRNA/JIMT-1 cells (i.e., ~ 2.5-fold decrease). The erlotinib IC50 value was diminished from 5.0 ± 1.0 μmol/L and 4.5 μmol/L in parental and negative control shRNA/JIMT-1 cells, respectively, to 1.5 ± 0.3 μmol/L (p < 0.05) in ATG8/LC3 shRNA/JIMT-1 cells (i.e., ~ 3-fold decrease). The lapatinib IC50 value dropped from 7.0 ± 1.0 μmol/L and 7.5 ± 1.0 μmol/L in parental and negative control shRNA/JIMT-1 cells, respectively, to 1.8 ± 0.3 μmol/L (p < 0.05) in ATG8/LC3 shRNA/JIMT-1 cells (i.e., ~ 4-fold decrease). Of note, when compared to parental and negative control shRNA/JIMT-1 cells, ATG8/LC3 shRNA/JIMT-1 cells did not increase their responses to cytotoxic therapies including the anthracycline doxorubicin, the pyrimidine analogue 5-fluorouracil, the DNA cross-linking agent cisplatin, the vinca alkaloid vinorelbine and the taxane paclitaxel ( Supplementary Fig. 3 ).
The anti-malarial chloroquine creates a trastuzumab-sensitive phenotype by blocking autophagic flux in breast cancer cells with primary resistance to trastuzumab
Given that cells engineered to express extremely low levels of key autophagy proteins involved in autophagosome formation gained some sensitivity to trastuzumab, we then decided to evaluate whether chloroquine's ability to inhibit the maturation of autophagosomes into degradative autophagolysosomes might create a phenocopy of shRNA ATG8/LC3-JIMT-1 cells in terms of trastuzumab responsiveness. Short-term treatment (6 h) with either chloroquine or trastuzumab alone increased the formation of autophagosomes, as shown by the increased number of LC3-positive granules ( Figure 3A , left panels). Notably, the combination of chloroquine with trastuzumab resulted in a significantly stronger accumulation of LC3-positive structures; this finding was consistent with a chloroquine-induced defect in autolysosomal degradation55. These findings also confirmed the autophagy-inducing effect of trastuzumab in trastuzumab-refractory breast cancer cells: if trastuzumab blocks autophagy, equal levels of LC3-positive cytoplasmic vesicles should be expected in cells treated with single-agent chloroquine and in cells treated with the combination of chloroquine and trastuzumab. Combination of chloroquine and trastuzumab for a longer period (48 h) induced a more robust accumulation of LC3 and p62/SQSTM1 co-aggregates ( Figure 3A , right panels).
Pharmacological ablation of the autophagy flux: Impact on the primary resistance to trastuzumab.
(A). Representative images of untreated, trastuzumab (Tzb)-, chloroquine and trastuzumab + chloroquine-treated JIMT-1 cell cultures (6 h) that were captured using different channels for LC3 (red) and Hoechst 33258 (blue). Right panels. Representative images of untreated, trastuzumab (Tzb)-, chloroquine and trastuzumab + chloroquine-treated JIMT-1 cell cultures (48 h) that were captured using different channels for LC3 (green), p62/SQSTM1 (red) and Hoechst 33258 (blue). Images were captured with a 20 × objective and merged on a BD Pathway™ 855 Bioimager System using BD Attovision™ software. Scale bar = 25 μm. (B). Top left. The metabolic status of JIMT-1 cells treated with graded concentrations of chloroquine was measured using MTT uptake assays and cell viability was expressed as% uptake of untreated control cells ( = 100% cell viability). Results are means (black squares) and 95% confidence intervals (bars) [n = 3]. Top right. The metabolic status of JIMT-1 cells treated with graded concentrations of trastuzumab in the absence or presence of different concentrations of chloroquine was measured using MTT uptake assays and cell viability was expressed as% uptake of untreated control cells ( = 100% cell viability). Results are means (symbols) and 95% confidence intervals (bars) of three independent experiments made in triplicate. Bottom. Figures indicate fold-increases in JIMT-1 cell kill at all drug concentrations over that expected for strict additivity between trastuzumab and chloroquine. (C). Confocal immunofluorescence images of annexin-V-Alexa 568 (red) and/or Hoechst 33258 staining in JIMT-1 cell cultures treated with trastuzumab in the absence or presence of chloroquine after 48 h. Scale bar = 25 μm. (D). The left side figure shows representative microphotographs of colonies formed on the plates of untreated, trastuzumab-treated, chloroquine-treated and trastuzumab + chloroquine-treated JIMT-1 cells, as specified. The right side bar graph shows the number of colonies formed after treatment of JIMT-1 cells with trastuzumab in the absence or presence of chloroquine, as specified. The error bar indicates the 95% confidence intervals for each mean (columns) generated from triplicate experiments. Statistically significant difference (one-factor ANOVA analysis) between experimental condition groups and control groups are shown.
To test whether chloroquine-induced inhibition of autophagy affected the survival of trastuzumab-refractory JIMT-1 cells, we first performed short-term (MTT-based) survival assays in cells treated with increasing concentrations of trastuzumab in the presence or absence of graded concentrations of chloroquine. Chloroquine concentrations up to 10 μmol/L did not significantly affect the viability of JIMT-1 cells when used in the absence of trastuzumab ( Figure 3B , top panels). As mentioned above, trastuzumab concentrations as high as 100 μg/mL failed to affect the viability of JIMT-1 cells. However, when used in combination, chloroquine and trastuzumab notably reduced cell viability in a synergistic manner. When used alone, trastuzumab IC50 values were not reached at the highest concentrations tested (i.e., IC50 > 1000 μg/mL trastuzumab; data not shown); however, less than 100 μg/mL trastuzumab was necessary to reduce cell viability by 50% in the presence of 10 μmol/mL chloroquine ( Figure 3B , top panels). The synergistic augmentation of cytotoxicity was not dose dependent because JIMT-1 cells showed higher fold increases in cytotoxicity when treated with 1 μmol/L chloroquine in the presence of trastuzumab (up to 45-fold with 25 μg/mL trastuzumab) than when treated with 10 μmol/L chloroquine in the presence of trastuzumab (up to 5-fold with 50 and 100 μg/mL trastuzumab) ( Figure 3B , bottom panels). These findings suggested that the incremental ability of chloroquine to sensitize tumor cells to trastuzumab becomes saturated; this saturation likely occurs because of the impossibility of further decreasing the bioactivity of the molecular point of convergence targeted by chloroquine and trastuzumab (see below). Importantly, accelerated cell death was observed in JIMT-1 cells co-treated with chloroquine and trastuzumab, as immunofluorescence microscopy of annexin V-stained cells showed a significant increase in the exposure of phosphatidylserine on the surface of apoptotic cells ( Figure 3C ); accordingly, we also observed a significant increase in the number of cells showing the homogeneous intense staining of the nucleus or nuclear fragments that is characteristic of apoptotic cells with nuclear condensation ( Figure 3C ). Given that chloroquine co-exposure apparently switches protective autophagy in trastuzumab-treated cells to apoptotic cell death, we hypothesized that when used together with trastuzumab, the ability of chloroquine to decrease cell viability should be more striking in long-term clonogenic assays. Accordingly, the more sensitive in vitro clonogenic assay revealed although that neither trastuzumab nor chloroquine altered the clonogenic capacity of JIMT-1 cells, combined treatment with chloroquine and trastuzumab markedly suppressed the frequency of surviving cells ( Figure 3D ).
To unambiguously confirm that autophagy inhibition accelerates trastuzumab-induced cell death in “autophagy-addicted” JIMT-1 cells, we monitored how the autophagic inhibitor chloroquine impacted in the appearance of active caspase-3, a major executioner caspase during the demolition phase of apoptosis. Virtually none of the cells in the trastuzumab-treated population exhibited apoptosis by the 48 h time point (1% with 100 μg/mL trastuzumab); indeed, the background signal of cleaved caspase-3 (green staining) within the cell nuclei of untreated cultures was not significantly altered in the presence of trastuzumab. Importantly, this activation was insignificant compared with the caspase-3 activity induced by chloroquine alone or by trastuzumab and chloroquine together ( Figure 4 ). In comparison to control conditions, we observed a dose-dependent increase in the percentage of cleaved caspase-3 JIMT-1 cells (calculated by dividing the total number of caspase 3-positive-cells by the total number of cells in the field of view) in response to graded concentrations of chloroquine (up to 37% with 20 μmol/L chloroquine). Importantly, the pharmacological inhibition of the autophagy pathway supra-additively accelerated the activation of the executioner caspase-3, which reached maximal levels of 60% in JIMT-1 cell cultures co-exposed to trastuzumab and chloroquine. To validate the specific functional role of key autophagy proteins such as ATG8/LC3, ATG5 and ATG12 in switching protective autophagy to apoptotic cell death in response to trastuzumab, we genetically inactivated autophagy by using the above-mentioned shRNA approach for their specific and stable inhibition in JIMT-1 cells. shRNAs to LC3B, ATG5 and ATG12 specifically suppressed the expression of the LC3B, ATG5 and ATG12 proteins and, more importantly, they resulted in trastuzumab-induced enhanced activation of caspase-3. Thus, the insignificant cleavage of caspase-3 in response to single-agent trastuzumab was notably increased by 4-, 9- and 23-fold in the absence of LC3B, ATG5 and ATG12, respectively ( Figure 4 ).
Pharmacological and genetic ablation of the autophagy pathway: Effect on caspase-3 activation in trastuzumab-treated cells.
(A). JIMT-1 cells were treated with trastuzumab, chloroquine or trastuzumab plus chloroquine as specified. 48 h after treatments, cells were processed for immunofluorescence staining with Hoechst 33258 (blue) and cleaved caspase-3 (green), which is indicative of apoptosis activation. F-actin filaments were labeled with Alexa Fluor® 555 phalloidin (red; #8953, Cell Signaling Technology, Inc., Danvers, MA, USA). Data show percentage of cells with positive cleaved caspase-3 cells calculated by dividing the total number of caspase-3-positive cells by the total number of cells in the field of view. (B). Top panel. shRNA-driven knockdown of autophagy-related autophagy proteins in JIMT-1 cells enhances trastuzumab-induced apoptotic cell death. Protein gel blot analyses of whole-cell lysates confirming knockdown of endogenous ATG8/LC3B, ATG5 and ATG12 autophagy-related proteins. The cropped blots portrayed are representative of three independent experiments. All gels were run under exact same experimental conditions for best comparison. β-actin was used as the internal control to confirm equal loading of lanes (β-actin [1] is the actual internal standard of the ATG12 gel; β-actin [2] is the actual internal standard of the ATG5 and LC3B gel). Bottom panel. Parental JIMT-1, negative control shRNA/JIMT-1, ATG8/LC3 shRNA/JIMT-1, ATG5 shRNA/JIMT-1 and ATG12 shRNA/JIMT-1 cells were treated with 100 μg/mL trastuzumab for 48 h and the percentage of caspase-3-positive cells was calculated as in (A). The results are presented as the means (columns) and 95% confidence intervals (bars) of two independent experiments performed in triplicate. Statistically significant differences comparing mean levels (one-factor ANOVA analysis) are shown. (n.s. not statistically significant).
Systemic chloroquine reduces tumor growth and sensitizes trastuzumab-resistant xenografts to trastuzumab
The effects of systemic chloroquine treatment on tumor growth were studied in vivo using a JIMT-1 xenograft animal model ( Figure 5 ). Compared with the untreated control group, which displayed mean tumor volumes of 940 ± 170 mm3, seven weeks of treatment with trastuzumab (5 mg/kg/week) failed to prevent tumor growth in animals xenografted with parental JIMT-1 cells, as the mean tumor volume reached volumes as high as 891 ± 135 mm3 in the presence of trastuzumab. However, compared with the mean xenograft tumor volume in both the untreated control and trastuzumab-treated groups, the mean tumor volume of the chloroquine-treated mice was significantly smaller (560 ± 54 mm3). Chloroquine did not cause any evident side effects in the treated animals. Remarkably, weekly co-treatment of the chloroquine-treated xenografts with trastuzumab resulted in a dramatic reduction in the mean tumor volume to 75 ± 54 mm3. These findings demonstrate that the trastuzumab-refractory JIMT-1 xenografts are sensitized to trastuzumab when the autophagic flux is pharmacologically ablated with chloroquine. Thus, whereas trastuzumab treatment reduced the tumor volume in autophagy-competent JIMT-1 xenografts by only 5% 49 days post-injection, combination treatment with trastuzumab and chloroquine reduced the tumor volume of JIMT-1 xenografts by an impressive 92% over the same treatment period ( Figure 5 ).
Blockade of autophagy by pharmacological inhibition: Impact on the efficacy of trastuzumab in vivo.
Left. Shown are the mean tumor volumes (±SD) in JIMT-1 xenograft-bearing nude mice following i.p. injection with trastuzumab (5 mg/kg/week) and/or chloroquine (20 ng/kg/daily) for nine weeks. Note that tumor volumes in the trastuzumab + chloroquine group were drastically reduced in a synergistic manner compared to the untreated control group and with the single agent treatments. Changes in Bax:Bcl-2 ratios in JIMT-1 xenografts treated with trastuzumab in the absence or presence of chloroquine are also shown. The results are presented as the means (columns) and 95% confidence intervals (bars) of two independent western blotting experiments following densitometric analyses of Bax and Bcl-2 proteins (calculations were based on the arbitrary values of the bands to show the Bax:Bcl-2 ratio in each tumor). Statistically significant difference (one-factor ANOVA analysis) between experimental condition groups and control groups are shown. Right. HER2 gene-amplified breast cancer cells with inherent (primary) resistance to trastuzumab are addicted to autophagy as the need for complementary bioenergetic resources in response to HER1/2-targeting drugs, which significantly affect cancer cells' addiction to certain fuel sources and metabolic pathways (“metabolic reprogramming”), is satisfied by the autophagy's ability to promote large-scale recycling of cytoplasmic macromolecules and organelles including mitochondria to regenerate energy and building blocks. In addition, protective autophagy in trastuzumab-refractory breast cancer cells appears to involve a rapid targeting of HER2 protein aggregates for degradation in the autophagolysosome. In this scenario, the concurrent combination of trastuzumab with pharmacologically (i.e., chloroquine)-induced autophagy dysfunction, with blockade of autophagosome and lysosome function and accumulation of autophagosomes and autophagy substrates, can lead to an aberrant accumulation of unmetabolized substrate with deleterious consequences (i.e., apoptotic cell death) in autophagy-addicted, trastuzumab-refractory breast cancer cells.
Since the balance between pro-apoptotic (Bax) and anti-apoptotic (Bcl-2) members of the Bcl-2 family of intracellular proteins serves as a rheostat to determine the susceptibility to apoptosis and because autophagy might actually protect tumour cells from apoptosis by modulating the expression of Bcl-2 family members and preventing the pro-apoptotic increase in Bax/Bcl-2 ratio, we investigated the role of chloroquine-induced blockade of autophagy flux in modulating the trastuzumab-unresponsive Bax/Bcl-2 ratio in trastuzumab-refractory JIMT-1 xenotumors. The autophagy inhibitor chloroquine alone did not result in a significant disturbance of Bax/Bcl-2 ratio ( Figure 5 ); importantly, the simultaneous combination of trastuzumab and chloroquine increased expression of Bax while reducing that of Bcl-2, which resulted in the synergistic increase in Bax/Bcl-2 ratio in JIMT-1 tumors ( Figure 5 ).
Discussion
Using a unique mechanistic platform involving an early explant of trastuzumab-refractory breast cancer cells that recapitulates the crucial biological features observed in HER2-positive breast cancer patients who progress rapidly after initial therapy with trastuzumab, we addressed the molecular nature of the trastuzumab therapeutic barriers ab initio and revealed for the first time that inherent resistance to trastuzumab and other HER2-targeted therapies can be molecularly explained in terms of protective autophagy. Because autophagy is deemed protective when blocking this process increases sensitivity to a stress stimulus, we concluded that protective autophagy causes the de novo resistance of breast cancer cells to HER2-targeted therapies. We propose that constitutive autophagic flux operates as an intrinsic cytoprotective mechanism against trastuzumab and provide proof-of-concept evidence for a novel drug combination of the HER2 inhibitor trastuzumab and the autophagy inhibitor chloroquine for more effective treatment of trastuzumab-refractory HER2 gene-amplified metastatic breast carcinomas.
Our findings confirmed the concept that HER2-overexpressing breast cancer cells can be addicted to autophagy for primary and secondary maintenance of the trastuzumab-resistant phenotype ( Figure 6 ). High levels of the lipidated form of LC3 accompanied by decreased p62/SQSTM1 cellular content, which are characteristic of increased autophagic flux, were previously observed during the development of acquired (secondary) resistance to trastuzumab44. Moreover, increased autophagy allowed cells to survive trastuzumab therapy, as silencing of ATG8/LC3 expression via transient transfection with siRNA was sufficient to significantly reduce proliferation rates in trastuzumab-adapted breast cancer cells and supra-additively interacted with trastuzumab to reverse acquired resistance. Breast cancer cells with intrinsic (primary) resistance to trastuzumab are also in a state of “autophagy addiction” because stable shRNA-driven silencing of autophagy-specific genes (i.e., ATG8/LC3) efficiently suppresses intrinsic refractoriness to trastuzumab and other HER-targeting drugs. Co-treatment with chloroquine significantly inhibited the autophagic flux, thereby maintaining high cellular contents of LC3-II and p62/SQSTM1 autophagic proteins, which were associated with synergistic cell death following co-treatment with chloroquine and trastuzumab. Although we recognize that the supra-additive interaction between chloroquine and trastuzumab may involve other molecular mechanisms, it should be noted that cells engineered to stably exhibit specific (shRNA-mediated) knockdown of autophagy-specific genes (ATG8/LC3) were sensitized to the growth-inhibitory and cytotoxic effects of HER2-targeted therapies. Because there are three different genes coding for LC3-like proteins (LC3A-C) and there four additional GABARAP-like proteins that likely compensate for a reduction in the protein levels of other mammalian ATG8s, it could be argued that specific knock-down of an essential gene for autophagy should be performed to unambiguously conclude that primary resistance to trastuzumab is an autophagy-dependent molecular phenomenon. Using low-density, autophagy-dedicated qRT-PCR-based platforms for monitoring primary resistance to HER2-targeted therapies by transcriptionally screening the autophagy interactome, we have recently revealed that the essential autophagy gene ATG12 –a positive regulator of autophagic vesicle formation- was the most differentially up-regulated genes in JIMT-1 cells and several other trastuzumab-unresponsive HER2-overexpressing breast cancer cells43. Lentiviral-delivered shRNA stable knockdown of the ATG12 gene fully suppressed the refractoriness of JIMT-1 cells to trastuzumab, erlotinib, gefitinib and lapatinib in vitro. Moreover, ATG12 silencing significantly reduced JIMT-1 tumor growth induced by subcutaneous injection in nude mice while the outgrowth of trastuzumab-unresponsive tumors was prevented completely when trastuzumab treatment was administered in an ATG12-silenced genetic background43. To further address the underlying mechanisms of the drastically enhanced trastuzumab-induced cell death occurring in response to pharmacological and genetic ablation of autophagy in JIMT-1 cells, we here monitored the processing of caspase-3. Because caspase-3 is cleaved in the late “execution” apoptotic phase and the combination of autophagy blockade and trastuzumab synergistically induced cleavage of caspase-3, it is reasonable to suggest that inhibition of the autophagy pathway results not only in earlier activation of executioner caspases (data not shown) but also accelerates this activation at later periods in trastuzumab-treated JIMT-1 cells. The fact that trastuzumab-induced apoptotic cell death is markedly enhanced by the specific inhibition of autophagy appear to confirm that protective autophagy plays a pivotal defensive role of certain subsets of HER2-positive breast cancer cells against HER2-targeted therapies.
Targeting the autophagy pathway to potentiate the efficacy of HER2-targeted drugs in breast cancer.
In HER2 gene-amplified breast carcinoma cells primarily responsive to trastuzumab, pharmacological blockade of HER2 signaling causes not only cell death but also autophagy, which acts as a survival mechanism that can be acquired during development of secondary resistance to trastuzumab (top panel). An enhanced basal autophagy may similarly confer primary (inherent) resistance to HER2-targeted drugs (bottom panel). The observations that anti-HER2 drugs alone induce autophagy44,59,60,61, coupled with the evidence that autophagy plays an active role in resistance to anti-HER2 agents, strongly suggest that additional studies should unambiguously define whether it represents a novel mechanism of resistance to anti-HER2 therapies in vivo. For instance, if trastuzumab-refractory patients have higher basal rates of autophagy than those that response well to trastuzumab-based therapies, the knowledge gained from such studies would enable clinicians to use the genetic status of key autophagy regulators (e.g., ATG1243) to predict therapeutic responses and tailor specific regimens (e.g., the FDA-approved anti-malarial agents chloroquine) to optimize clinical outcomes for HER2 gene-amplified breast cancer patients. Blocking HER2 and autophagy simultaneously might overcome both inherent and acquired resistance and restore sensitivity of HER2 gene-amplified breast cancer cells to HER2-targeted therapies including trastuzumab.
When used in combination with trastuzumab, chloroquine treatment was sufficient to create a cellular state that was sensitive to the growth-inhibitory activity of trastuzumab. The ability of chloroquine to overcome or circumvent primary resistance to trastuzumab was more striking in long-term clonogenic assays than in short-term MTT assays, indicating that the combinatorial treatment not only alters the kinetics of cell survival/cell death but actually decreases the number of cells that can survive the combination treatment and that can subsequently proliferate and form colonies once the co-treatment is ceased. Indeed, when chloroquine impeded the autophagic resolution of the accumulation of autophagolysosomes generated in the presence of trastuzumab, cells committed to die by apoptosis. Moreover, combination treatment with trastuzumab and chloroquine radically suppressed tumor growth by > 90% in a trastuzumab-refractory breast tumor xenograft model, further underscoring the vulnerability of trastuzumab-refractory cells to any impairment in autophagic flux. This finding also suggests that several other mechanisms may converge to synergistically potentiate the molecular addiction to autophagy of HER2-positive subgroups of breast carcinomas exhibiting primary resistance to HER2-targeted therapies. For example, trastuzumab-refractory JIMT-1 cells constitutively exhibit low levels of AKT activity in the absence of phosphatase and tensin homologue (PTEN), a molecular combination that cooperates to promote the autophagic survival of drug-resistant tumor cells but sensitizes them to lysosomotropic agents55,56. Moreover, recent approaches in our laboratory have exposed a new scenario in which the relative presence of “autophagic CSCs” may specifically regulate the refractoriness to trastuzumab in certain subsets of HER2 gene-amplified breast carcinomas; CD44+CD24-/low mesenchymal subpopulations are intrinsically unresponsive to trastuzumab; the presence or absence of the CD44+CD24-/low mesenchymal immunophenotype is closely related to the inherent refractoriness or sensitivity of HER2 gene-amplified breast carcinoma cells to trastuzumab, respectively; and the impairment of autophagic flux by genetic means impedes the ontogeny of generating the CSC-like CD44+CD24-/low mesenchymal phenotype by preventing the full acquisition of a post-EMT status, thus decreasing the number of trastuzumab-unresponsive cells bearing the CD44+CD24-/low cell surface phenotype35,36,54. It is noteworthy that, while treatment with trastuzumab failed to impede the propensity of breast cancer stem cells to form multicellular “microtumors” in non-adherent and non-differentiating conditions (i.e., “mammospheres”)29, co-treatment with chloroquine and trastuzumab had a profound inhibitory effect on mammosphere growth (data not shown). Altogether, these findings may suggest that the inhibition of autophagic flux can markedly increase the inhibitory effects of trastuzumab on trastuzumab-refractory cells with a CSC-like phenotype.
The notion that targeting autophagy when it promotes the survival of cancer cells under stress would achieve selective antitumor efficacy became more evident when assessing the nature of the tumoricidal interaction between chloroquine and trastuzumab in vivo. It appears that, likely by inducing further autophagic flux, treatment with trastuzumab enhances the dependence of the tumors on autophagy for survival and growth. This enhanced dependence on autophagy underlies the enhanced antitumor activity of co-administered chloroquine and trastuzumab against trastuzumab-refractory HER2 gene-amplified breast cancer cells. For the first time, our findings confirm this scenario in vivo, as co-treatment with chloroquine and trastuzumab was dramatically more effective in reducing the tumor growth of trastuzumab-refractory xenograft tissues implanted in mice than treatment with each agent alone. The molecular mechanisms underlying these observations might include the interference of protective autophagy with the trastuzumab-mediated dysregulation of the balance between pro-apoptotic and anti-apoptotic members of Bcl-2 family, as suggested by our data that pharmacological blockade of autophagy increases Bax/Bcl-2 ration in trastuzumab-treated JIMT-1 xenotumors. Because increase in Bax:Bcl-2 ratio could increase mitochondrial release of pro-apoptotic molecules to trigger activation of downstream processes of apoptosis, the drastic reduction in the tumor volume of trastuzumab-refractory JIMT-1 xenotumors occurring in the concomitant presence of trastuzumab and chloroquine might be explained in terms not only of reduced cell proliferation but also induction of cell death. Considering reports of failing chloroquine efficacy and disappointing results of various combination chloroquine/anti-cancer therapy regimens22, our instructive, insightful observations strongly suggest that the use of improved versions of chloroquine may prove to have a major impact in combination with HER2-targeted therapies, including trastuzumab.
Cancer is increasingly recognized as a metabolic disease57,58. Therefore, similar to other capacities that define the malignant process (the so-called “cancer hallmarks”), autophagy can no longer be viewed simply as a secondary phenotype programmed by oncogenes and tumor suppressor genes. As such, the clinical evaluation of autophagy-targeting drugs (e.g., chloroquine or hydroxychloroquine) should include biomarker-based pre-selection (e.g., autophagy addiction) within appropriate patient populations (e.g., trastuzumab-refractory, HER2 gene-amplified breast cancer patients) to ensure that molecular and clinical responses directly relate to proper target engagement (i.e., autophagy flux inhibition) in the tumor. Only then will we be able to develop new, more effective anti-cancer treatment modalities by combining autophagy inhibitors with molecularly targeted drugs that, although directed against specific cellular targets, notably impact the strict metabolic reprogramming of breast cancer cells.
Methods
Drugs
The EGFR (HER1)-Tyrosine Kinase Inhibitor (TKI) gefitinib (ZD1839; Iressa) was kindly provided by AstraZeneca (AstraZeneca PLC Headquarters, 15 Stanhope Gate, W1K 1 LN, London, UK). The EGFR (HER1) TKI erlotinib (Tarceva®) was a kind gift from Roche Pharmaceuticals (Neuilly sur Seine, France). The dual HER1/HER2-TKI1 lapatinib (GW572016; Tykerb®) was kindly provided by GlaxoSmithKline (GSK), Corporate Environment, Health & Safety (Brentford, Middlesex TW8 9GS, UK). Stock solutions of gefitinib, erlotinib and lapatinib (10 mmol/L) were prepared in DMSO and stored in aliquots in the dark at −20°C until use. Trastuzumab (Herceptin®) was kindly provided by Hospital Universitari de Girona Dr. Josep Trueta Pharmacy (Girona, Spain). Trastuzumab was solubilized in bacteriostatic water containing 1.1% benzyl alcohol (stock solution at 21 mg/mL) for injection, stored at 4°C and used within 1 month. For experimental use, all agents were prepared fresh from stock solutions and were diluted with cell growth medium. The control cells were cultured in media containing concentrations (v/v) of drugs identical to those used for the experimental cells. The vehicle solutions had no noticeable influence on the proliferation of the experimental cells.
Materials
Bafilomycin A1 and chloroquine were purchased from Sigma-Aldrich (St. Louis, MO, USA) and reconstituted in dimethyl sulfoxide (DMSO) and phosphate-buffered saline (PBS), respectively. Rabbit anti-light-chain 3 (LC3) polyclonal antibody was purchased from MBL International Corporation (Woburn, MA, USA; PD014). Alternatively, we employed an anti-LC3 rabbit polyclonal antibody purchased from Novus Biologicals (NB100-2331). Mouse anti-SQSTM1/p62 monoclonal antibody was purchased from Abcam plc. (Cambridge, UK; ab56416). Cleaved caspase-3 (Asp175) (5A1E) rabbit monoclonal antibody (#9664) was purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Anti-ATG5 rabbit polyclonal antibody was purchased from Novus Biologicals (NB110-53818). Anti-ATG12 rabbit polyclonal antibody (human specific; #2010) was purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). Anti-β-actin mouse monoclonal antibody (C4, #sc-47778) was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA).
Cell lines and culture conditions
JIMT-1 cells were obtained from the German Collection of Microorganisms and were routinely grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS; Bio-Whittaker, Inc.), 1% L-glutamine, 1% sodium pyruvate, 50 U/mL penicillin and 50 μg/mL streptomycin. The cells were maintained at 37°C in a humidified atmosphere with 5% CO2. SKBR3 breast cancer cells were obtained from the American Type Culture Collection (ATCC) and were routinely grown in Improved MEM (IMEM; Biosource International) supplemented with 10% FBS and 1% L-glutamine, 1% sodium pyruvate, 50 U/mL penicillin and 50 μg/mL streptomycin. The cells were maintained at 37°C in a humidified atmosphere of 95% air and 5% CO2. The cells were screened periodically for Mycoplasma contamination.
Immunoblotting procedures
The cells were washed twice with cold PBS and then lysed in buffer (20 mmol/L Tris [pH 7.5], 150 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1% Triton® X-100, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerolphosphate, 1 mmol/L Na3VO4, 1 μg/mL leupeptin, 1 mmol/L phenylmethylsulfonylfluoride and complete protease inhibitor cocktail [Sigma-Chemicals, St. Louis, MO, USA]) for 30 minutes on ice. The lysates were cleared by centrifugation in an Eppendorf tube (15 minutes at 14,000 × g, 4°C). Protein content was determined against a standardized control using the Pierce Protein Assay Kit (Rockford, IL, USA). Equal amounts of protein (i.e., 50 μg) were resuspended in 5 × Laemmli sample buffer (10 min at 70°C), resolved by electrophoresis on 10% SDS-PAGE and transferred onto nitrocellulose membranes. Non-specific binding to the nitrocellulose filter paper was minimized by blocking for 1 h at RT with TBS-T buffer [25 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 0.05% Tween 20] containing 5% (w/v) nonfat dry milk. The treated filters were washed in TBS-T and then incubated overnight with primary antibodies, as specified, in 5% w/v BSA, 1 × TBS-T buffer, 0.1% Tween-20 at 4°C with gentle shaking. The membranes were washed with TBS-T, horseradish peroxidase-conjugated secondary anti-mouse/rabbit IgGs in TBS-T was added for 1 h and immunoreactive bands were detected using a chemiluminescence reagent (Pierce, Rockford, IL, USA). The experiments involving immunoblotting procedures were repeated at least three times and the blots were re-probed with an antibody against β-actin to control for protein loading and transfer. The densitometric values of protein bands were quantified using the Scion Image software (Scion Corporation, Frederick, MD, USA).
Immunofluorescence staining and high-content confocal imaging
The cells were seeded at approximately 5,000 cells/well in 96-well clear bottom imaging tissue culture plates (Becton Dickinson Biosciences; San Jose, CA, USA) optimized for automated imaging applications. Triton® X-100 permeabilization and blocking, primary antibody staining (1:50 dilution), secondary antibody staining using Alexa Fluor® 488/594 goat anti-rabbit/mouse IgGs (Invitrogen, Molecular Probes, Eugene, Oregon, USA) and counterstaining (using Hoechst 33258; Invitrogen) were performed following BD Biosciences protocols. Images were captured in different channels for Alexa Fluor® 488 (pseudo-colored green), Alexa Fluor® 594 (pseudo-colored red) and Hoechst 33258 (pseudo-colored blue) on a BD Pathway™ 855 Bioimager System (Becton Dickinson Biosciences, San Jose, California, USA) using 20 × or 40 × objectives (NA 075 Olympus). Merged images were obtained according to the Recommended Assay Procedure using BD Attovision™ software.
GFP-LC3 overexpression
SKBR3 and JIMT-1 cells were plated at a density of 2 × 105 cells on glass coverslips in six-well plates and cultured up to 70% confluence. Cells were transfected with GFP-LC3 plasmid DNA (2 μg/mL of plasmid DNA in each well used) for 36 h and then cultured in regular medium in the absence or presence of trastuzumab for 24 h. Transfection was carried out with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) as per manufacturer's recommendations. The cells were fixed and the nuclei were stained with Hoechst 33258. The fluorescence of GFP-LC3 was observed under a confocal fluorescence microscope. Cells containing five or more GFP-LC3 dots were defined as autophagy-positive cells.
Lentiviral transduction
Pre-packaged lentiviral particles that either encoded a non-targeting shRNA (negative shRNA; sc-108080) or sequences specifically targeting the human ATG8/LC3 gene (sc-43390-V) were purchased from a commercial provider (Santa Cruz Biotechnology). For viral infection of JIMT-1 cells, the regular medium was replaced with culture medium containing 5 μg/mL polybrene (Santa Cruz Biotechnology; sc-124220). JIMT-1 cells were then exposed to lentiviruses for 48 h. Because the lentiviral shRNA particles also encode a puromycin resistance gene for transduction selection, the cells were then washed and grown in culture medium containing 10 μg/mL puromycin dihydrochloride (Sigma; P9620) for an additional 72 h. The JIMT-1 cells were allowed to recover and proliferate for at least 1 week before any experimental procedures and the cells were then analyzed. To monitor the lentiviral transduction efficiency and transgene expression for the duration of the experiment, we incubated additional subsets of JIMT-1 cells with lentiviral particles encoding a green fluorescence protein (GFP) reporter (sc-108084). Transduction efficiency (> 90%) was calculated as the ratio of the number of GFP-positive cells to the total number of cells from five random visual fields from three independent culture experiments.
Metabolic status assessment (MTT-based cell viability assays)
Cell viability was determined using a standard colorimetric MTT (3-4,5-dimethylthiazol-2-yl-2, 5-diphenyl-tetrazolium bromide) reduction assay. Exponentially growing cells were harvested by trypsinization, seeded at a concentration of ~ 2.5 × 103 cells/200 μL/well into 96-well plates and allowed to attach overnight. The medium was then removed and fresh medium along with various concentrations of HER1/2-targeting drugs was added to the cultures as specified. Control cells without drugs were cultured in parallel using the same conditions with comparable media changes. The drugs were not renewed during the entire period of cell exposure. Following treatment (5 days), the medium was removed and replaced with fresh, drug-free medium (100 μL/well) and MTT (5 mg/mL in PBS) was added to each well at a 1:10 dilution. After incubation for 2–3 h at 37°C, the supernatants were carefully aspirated, 100 μL DMSO was added to each well and the plates were agitated to dissolve the crystal product. The optical density (OD) was measured at 570 nm in a multi-well plate reader (Model Anthos Labtec 2010 1.7 reader). The cell viability effects resulting from the exposure of cells to HER1/2-targeting drugs were analyzed as percentages of the control cell absorbances, which were obtained from control wells treated with the appropriate concentrations of the agents' vehicles, which were processed simultaneously. For each treatment, cell viability was evaluated as a percentage using the following equation: (OD570 of treated sample/OD570 of untreated sample) × 100. Breast cancer cell sensitivity to HER1/2-targeting drugs was expressed in terms of the concentration of drug required to decrease cell viability by 50% (IC50 value). Because the fraction of cells surviving after treatment was determined by comparing the absorbance in treated samples to that in control samples, the IC50 value was defined as the concentration of HER1/2-targeting drug that produced a 50% reduction in the absorbance of treated samples when compared to the control absorbance (by interpolation). The degree of sensitization to HER1/2-targeting drugs by shRNA-driven silencing of ATG8/LC3 was evaluated by dividing the IC50 values of the parental JIMT-1 cells by those obtained in control shRNA/JIMT-1 cells or ATG8/LC3 shRNA/JIMT-1 cells.
Clonogenic assays
JIMT-1 cells were cultured in six-well plates at a density of 1000 cells per well (in triplicate) and incubated for 18 h to allow for attachment. The cells were then treated with regular medium in the absence or presence of 100 μg/mL trastuzumab, 1 μmol/L chloroquine, or the trastuzumab + chloroquine combination for 3days; the cells were allowed to grow in a drug-free medium for an additional 10 days before the colonies were stained with crystal violet.
Xenograft studies
To produce xenografts, approximately 5 × 106 parental JIMT-1 cells were injected subcutaneously into the dorsal flanks of female athymic nude mice (four to five weeks old; 23 to 25 g; Harlan Laboratories, France). The animals were randomized into four groups, with five animals in each group: control (vehicle-treated), trastuzumab-treated, chloroquine-treated and [trastuzumab + chloroquine]-treated. Trastuzumab (5 mg/kg) was administered intraperitoneally (i.p.) once per week. Chloroquine (20 mg/kg) was administered i.p. once daily. The mice were weighed once per week after dosing, the tumor sizes were measured daily with electronic calipers and the tumor volumes were calculated using the following formula: volume (mm3) = length × width2 × 0.5. Protein samples were extracted following the lysis of control and drug-treated tumor tissues, quantitated spectrophotometrically, denatured in boiling water for 5 min and loaded onto the SDS-polyacrylamide gels (Bio-Rad, Hercules, CAS, USA). All proteins were resolved by electrophoresis and then electroblotted to the membranes. The blots were re-probed with an antibody against β-actin to control for protein loading and transfer. Bax and Bcl-2 immunoreactive bands were detected using a chemiluminescence reagent (Pierce, Rockford, IL, USA) and the densitometric values of protein bands were quantified using the Scion Image software (Scion Corporation, Frederick, MD, USA). The experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the Institut d'Investigació Biomèdica de Bellvitge (IDIBELL; Animal Use Protocol #6302 authorized by the Animal Experimental Commission of the Catalan Government, Barcelona, Spain).
References
Chong, C. R. & Sullivan, D. J. Jr. New uses for old drugs. Nature 448, 645–646 (2007).
Ashburn, T. T. & Thor, K. B. Drug repositioning: identifying and developing new uses for existing drugs. Nat Rev Drug Discov 3, 673–683 (2004).
Reichert, J. M. Trends in development and approval times for new therapeutics in the United States. Nat Rev Drug Discov 2, 695–702 (2003).
Wiesner, J., Ortmann, R., Jomaa, H. & Schlitzer, M. New antimalarial drugs. Angew Chem Int Ed Engl 42, 5274–5293 (2003).
Breckenridge, A. M. & Winstanley, P. A. Clinical pharmacology and malaria. Ann Trop Med Parasitol 91, 727–733 (1997).
Solomon, V. R. & Lee, H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur J Pharmacol 625, 220–233 (2009).
He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 43, 67–93 (2009).
Klionsky, D. J. & Emr, S. D. Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721 (2000).
Tanida, I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal 14, 2201–2214 (2011).
Pyo, J. O., Nah, J. & Jung, Y. K. Molecules and their functions in autophagy. Exp Mol Med 44, 73–80 (2012).
Yang, Z. J., Chee, C. E., Huang, S. & Sinicrope, F. A. The role of autophagy in cancer: therapeutic implications. Mol Cancer Ther 10, 1533–1541 (2011).
Giansanti, V. et al. Killing of tumor cells: a drama in two acts. Biochem Pharmacol 82, 1304–1310 (2011).
Janku, F., McConkey, D. J., Hong, D. S. & Kurzrock, R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol 8, 528–539 (2011).
Amaravadi, R. K. et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res 17, 654–666 (2011).
Mancias, J. D. & Kimmelman, A. C. Targeting autophagy addiction in cancer. Oncotarget 2, 1302–1306 (2011).
Boya, P. et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 25, 1025–1040 (2005).
Hu, C., Solomon, V. R., Ulibarri, G. & Lee, H. The efficacy and selectivity of tumor cell killing by Akt inhibitors are substantially increased by chloroquine. Bioorg Med Chem 16, 7888–7893 (2008).
Jiang, P. D. et al. Antitumor and antimetastatic activities of chloroquine diphosphate in a murine model of breast cancer. Biomed Pharmacother 64, 609–614 (2010).
Degtyarev, M. et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol 183, 101–116 (2008).
Fan, Q. W. et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal 3, ra81 (2010).
Maycotte, P. et al. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy 8, 200–212 (2012).
Gorski, S. M., Ries, J. & Lum, J. J. Targeting autophagy: The Achilles' heel of cancer. Autophagy 8, 1279–1280 (2012).
McAfee, Q. et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A 109, 8253–8258 (2012).
Amaravadi, R. K. & Winkler, J. D. Lys05: A new lysosomal autophagy inhibitor. Autophagy 8, 1383–1384 (2012).
Liu, J. et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147, 223–234 (2011).
Tanner, M. et al. Characterization of a novel cell line established from a patient with Herceptin-resistant breast cancer. Mol Cancer Ther 3, 1585–1592 (2004).
Nagy, P. et al. Decreased accessibility and lack of activation of ErbB2 in JIMT-1, a herceptin-resistant, MUC4-expressing breast cancer cell line. Cancer Res 65, 473–482 (2005).
Köninki, K. et al. Multiple molecular mechanisms underlying trastuzumab and lapatinib resistance in JIMT-1 breast cancer cells. Cancer Lett 294, 211–219 (2010).
Vazquez-Martin, A., Oliveras-Ferraros, C., Del Barco, S., Martin-Castillo, B. & Menendez, J. A. The anti-diabetic drug metformin suppresses self-renewal and proliferation of trastuzumab-resistant tumor-initiating breast cancer stem cells. Breast Cancer Res Treat 126, 355–364 (2011).
Damiano, V. et al. A novel toll-like receptor 9 agonist cooperates with trastuzumab in trastuzumab-resistant breast tumors through multiple mechanisms of action. Clin Cancer Res 15, 6921–6930 (2009).
Oliveras-Ferraros, C. et al. Dynamic emergence of the mesenchymal CD44(pos)CD24(neg/low) phenotype in HER2-gene amplified breast cancer cells with de novo resistance to trastuzumab (Herceptin). Biochem Biophys Res Commun 397, 27–33 (2010).
Oliveras-Ferraros, C. et al. Pathway-focused proteomic signatures in HER2-overexpressing breast cancer with a basal-like phenotype: new insights into de novo resistance to trastuzumab (Herceptin). Int J Oncol 37, 669–678 (2010).
Valabrega, G. et al. HER2-positive breast cancer cells resistant to trastuzumab and lapatinib lose reliance upon HER2 and are sensitive to the multitargeted kinase inhibitor sorafenib. Breast Cancer Res Treat 130, 29–40 (2011).
Oliveras-Ferraros, C. et al. Inhibitor of Apoptosis (IAP) survivin is indispensable for survival of HER2 gene-amplified breast cancer cells with primary resistance to HER1/2-targeted therapies. Biochem Biophys Res Commun 407, 412–419 (2011).
Cufi, S. et al. Metformin-induced preferential killing of breast cancer initiating CD44+CD24-/low cells is sufficient to overcome primary resistance to trastuzumab in HER2+ human breast cancer xenografts. Oncotarget 3, 395–398 (2012).
Oliveras-Ferraros, C. et al. Epithelial-to-mesenchymal transition (EMT) confers primary resistance to trastuzumab (Herceptin). Cell Cycle 11, 4020–4032 (2012).
Barok, M. et al. Trastuzumab causes antibody-dependent cellular cytotoxicity-mediated growth inhibition of submacroscopic JIMT-1 breast cancer xenografts despite intrinsic drug resistance. Mol Cancer Ther 6, 2065–2072 (2007).
Ahtiainen, L. et al. Defects in innate immunity render breast cancer initiating cells permissive to oncolytic adenovirus. PLoS One 5, e13859 (2010).
Rubinsztein, D. C., Gestwicki, J. E., Murphy, L. O. & Klionsky, D. J. Potential therapeutic applications of autophagy. Nat Rev Drug Discov 6, 304–312 (2007).
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy Research. Cell 140, 313–326 (2010).
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 (2012).
Sarkar, S., Davies, J. E., Huang, Z., Tunnacliffe, A. & Rubinsztein, D. C. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem 282, 5641–5652 (2007).
Cufí, S. et al. Autophagy-related gene 12 (ATG12) is a novel determinant of primary resistance to HER2-targeted therapies: Utility of transcriptome analysis of the autophagy interactome to guide breast cancer treatment. Oncotarget 3, 1600–1614 (2012).
Vazquez-Martin, A., Oliveras-Ferraros, C. & Menendez, J. A. Autophagy facilitates the development of breast cancer resistance to the anti-HER2 monoclonal antibody trastuzumab. PLoS One 4, e6251 (2009).
Bjørkøy, G. et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171, 603–614 (2005).
Bjørkøy, G., Lamark, T. & Johansen, T. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2, 138–139 (2006).
Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282, 24131–24145 (2007).
Ichimura, Y., Kominami, E., Tanaka, K. & Komatsu, M. Selective turnover of p62/A170/SQSTM1 by autophagy. Autophagy 4, 1063–1066 (2008).
Shvets, E. & Elazar, Z. Autophagy-independent incorporation of GFP-LC3 into protein aggregates is dependent on its interaction with p62/SQSTM1. Autophagy 4, 1054–1056 (2008).
Yorimitsu, T. & Klionsky, D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ 12, 1542–1552 (2005).
Xie, Z. & Klionsky, D. J. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 9, 1102–1109 (2007).
Geng, J. & Klionsky, D. J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep 9, 859–864 (2008).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27, 107–132 (2011).
Cufí, S., Vazquez-Martin, A., Oliveras-Ferraros, C., Martin-Castillo, B., Vellon, L. & Menendez, J. A. Autophagy positively regulates the CD44(+) CD24(-/low) breast cancer stem-like phenotype. Cell Cycle 10, 3871–3885 (2011).
Degtyarev, M. et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol 183, 101–116 (2008).
Fan, Q. W. et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal 3, ra81 (2010).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Ward, P. S. & Thompson, C. B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297–308 (2012).
Cheng, Y., Li, H., Ren, X., Niu, T., Hait, W. N. & Yang, J. Cytoprotective effect of the elongation factor-2 kinase-mediated autophagy in breast cancer cells subjected to growth factor inhibition. PLoS One 5, e9715 (2010).
Huang, H. L. et al. Lapatinib induces autophagy, apoptosis and megakaryocytic differentiation in chronic myelogenous leukemia K562 cells. PLoS One 6, e29014 (2011).
Han, W. et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS One 6, e18691 (2011).
Acknowledgements
This work was financially supported by the Instituto de Salud Carlos III (Ministerio de Sanidad y Consumo, Fondo de Investigación Sanitaria (FIS), Spain, grants CP05-00090, PI06-0778 and RD06-0020-0028), the Fundación Científica de la Asociación Española Contra el Cáncer (AECC, Spain) and the Ministerio de Ciencia e Innovación (SAF2009-11579 and SAF2012-389134, Plan Nacional de I+D+ I, MICINN, Spain). Alejandro Vazquez-Martin received a Sara Borrell post-doctoral contract (CD08/00283, Ministerio de Sanidad y Consumo, Fondo de Investigación Sanitaria -FIS-, Spain). Sílvia Cufí received a research fellowship (Formación de Personal Investigador, FPI) from the Ministerio de Ciencia e Innovación (MICINN, Spain). The funders had no role in study design, data Collection and analysis, decisión to publish, or prepraration of the manuscript.
Author information
Authors and Affiliations
Contributions
S.C., A.V.M. and C.O.-F. performed the experiments, analyzed the data and prepared the figures; B.C.F., E.C. and E.L.-B. performed the experiments and analyzed the data; J.J. and B.M.C. designed the experiments and analyzed the data; and J.A.M. developed the hypothesis, designed the study, analyzed the data and wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Figures
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Cufí, S., Vazquez-Martin, A., Oliveras-Ferraros, C. et al. The anti-malarial chloroquine overcomes Primary resistance and restores sensitivity to Trastuzumab in HER2-positive breast cancer. Sci Rep 3, 2469 (2013). https://doi.org/10.1038/srep02469
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02469
This article is cited by
-
Advances in the study of autophagy in breast cancer
Breast Cancer (2024)
-
Exosomal Linc00969 induces trastuzumab resistance in breast cancer by increasing HER-2 protein expression and mRNA stability by binding to HUR
Breast Cancer Research (2023)
-
Gasdermin B over-expression modulates HER2-targeted therapy resistance by inducing protective autophagy through Rab7 activation
Journal of Experimental & Clinical Cancer Research (2022)
-
Long non-coding RNA LINC01559 exerts oncogenic role via enhancing autophagy in lung adenocarcinoma
Cancer Cell International (2021)
-
Targeting autophagy to overcome drug resistance: further developments
Journal of Hematology & Oncology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.