Abstract
The mobility of damaged chromatin regions in the nucleus may affect the probability of mis-repair. In this work, live-cell observation and distance tracking of GFP-tagged DNA damage response protein MDC1 was used to study the random-walk behaviour of chromatin domains containing radiation-induced DNA double-strand breaks (DSB). Our measurements indicate a subdiffusion-type random walk process with similar time dependence for isolated and clustered DSBs that were induced by 20 MeV proton or 43 MeV carbon ion micro-irradiation. As compared to normal diffusion, subdiffusion enhances the probability that both ends of a DSB meet, thus promoting high efficiency DNA repair. It also limits their probability of long-range movements and thus lowers the probability of mis-rejoining and chromosome aberrations.
Similar content being viewed by others
Introduction
DSB are a major threat for genomic integrity. However, some pathways for DSB repair can mis-rejoin DNA ends from different break sites, thus creating chromosome aberrations. The influence of initial distance between DSB sites on the probability of mis-rejoining is under debate, as well as the possibility of mechanisms bringing DSB sites into closer vicinity during damage processing1. Estimates on break interaction distances based on biophysical modelling vary between 100 nm and several μm2,3,4. Simulation models for DNA repair based on normal diffusion of ends tend to overestimate the fraction of unrejoined DSB after long repair times5.
Experimental data on the mobility of break sites in mammalian chromosomes are inconclusive. Mostly, they rely on labelling the chromatin region containing the DSB by visualizing specific chromatin modifications known to surround the breaks site, such as γ-H2AX, or proteins known to bind chromatin in the vicinity of break sites, such as MDC1. The labelled chromatin regions microscopically appear as ionizing radiation-induced foci (IRIF). Analyzing fixed cells, inferences on mobility are made by investigating the development of foci number and size over time, or their position with regard to the sites of damage induction6,7,8,9,10,11,12. In live-cell analysis, also the behaviour of individual foci over time can be observed and several authors measured trajectories in relation to the origin (single particle tracking13,14). In this case, thorough correction for translational and rotational movement of the whole nucleus, as well as its deformation, is paramount, since otherwise the measured mean squared displacements MSD(Δt) = <r2(Δt)> reflect movement of the nucleus rather than that of the foci within the nucleus.
A plot of MSD versus observation time allows determining if the particle observed moves via normal diffusion, in which case the MSD is linearly related to time and a diffusion coefficient D can be determined15,16. Assuming a linear dependence of MSD on time, recently a large diffusion coefficient was determined13 which was difficult to reconcile with the long-term stability of microscopic foci patterns after irradiation with ions in geometric patterns9. In addition, others reported for regions of undamaged chromatin that mobility is best described by subdiffusion (reviewed by17). In subdiffusion, MSD does not follow linear time dependence, but obeys

with Dα being a generalized diffusion coefficient and α being the anomality parameter quantifying the deviation from normal diffusion. In the case of subdiffusion, α < 1, while in normal diffusion, α = 1. Γ represents the Gamma function and d is the dimension of the analyzed space.
To characterize the mobility of damaged chromatin regions, we chose to investigate distance changes between two IRIF over time rather than the MSD, using the standard deviation of the distance changes σ2(Δl). While measuring distance changes has been introduced by others18,19 due to the obvious advantage over single-particle tracking in case of any translational and rotational movements of the nucleus, our method using the σ2(Δl) is even largely invariant in the case of additional changes of nucleus size.
Results
IRIF mobility follows a subdiffusion process
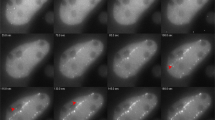
U2OS cells expressing GFP-tagged repair protein MDC1 were irradiated in a matrix pattern with single carbon (12C) ions per point. The distances li of adjacent MDC1 foci pairs numbered by i were monitored for several hours post-irradiation (Fig. 1). The distance changes Δli(Δt) during different time intervals Δt (i.e. Δli(Δt) = li(tj + Δt) − li(tj) for all time points tj of the time series) were determined.
U2OS cell irradiated with carbon ions in a 5 × 5 μm2 matrix pattern, visible under the fluorescence microscope by GFP-tagging of DNA repair protein MDC1 which accumulates at the radiation induced double-strand breaks (“foci”).
The distance li between neighbouring foci is recorded at several time points tj for analyzing the distance changes Δli over different time intervals Δt.
For each cell, we determined the standard deviation σ2(Δl(Δt)) of the changes in foci distances Δli(Δt) between neighbouring MDC1 foci as a measure for the underlying random walk process (see SI). Then the time-ensemble average of the “neighbouring” σ2(Δt) from all analyzed carbon irradiated cells was calculated for each Δt and plotted versus Δt in a double-logarithmic plot (Fig. 2, filled squares). A linear increase of log(σ2) with log(Δt) is evident, which demonstrates a power-law dependence of σ2 on Δt (cf. Supplementary equation (S1)):

We obtain α = 0.49 ± 0.05 (SEM) and D0.49 = (3.7 ± 0.5) × 10−4 μm2/s0.49 for the next neighbour foci (l ≈ 5 μm) of the carbon irradiated cells (R2 = 0.97, reduced χ2 = 1.6). The value obtained for α significantly differs from α = 1 demonstrating that the mobility of the IRIFs follows a subdiffusion process20. Our data do not support models based on constrained diffusion21, since we did not see indications for a limit to the radius within which the IRIF can diffuse, in spite of the long time range (up to 104 s) observed. In addition, the best least-square fits of constrained diffusion19 (R2 = 0.91, reduced χ2 = 5.0) or a combination of constrained diffusion and normal diffusion of the confinement region (R2 = 0.95, reduced χ2 = 2.8) did not fit the experimental data (Supplementary Fig. S2).
Double-logarithmic plot of the squared standard deviations σ2 (±SEM) of the distance changes Δl(Δt) between neighbouring MDC1 foci (filled squares) and second-next neighbours (open circles) in the nuclei of cells irradiated with carbon ions (657 foci pairs from 58 cells, 30–60 time frames per sample).
The data are fitted with the power-law function (equation (2)).
To examine the influence of the initial distances on the determined dynamics, the cells were additionally analyzed using the distances between second next foci neighbours (l ≈ 10 μm). As shown in Fig. 2 (open circles), the σ2 analyzed from both data sets are very similar for Δt < 180 s, but increase faster in the case of second next neighbours at Δt > 180 s, hinting at non-corrected additional relative foci movements which we attribute to gross morphological deformations of cell nuclei. We determined α = 0.58 ± 0.05 (SEM) and D0.58 = (3.1 ± 0.5) × 10−4 μm2/s0.58.
Comparison between time-series recorded early (3–10 min) and late (80–100 min) after irradiation shows that the subdiffusion of the MDC1 foci is time-invariant, justifying the used method of ensemble- and time-averaging as it can be done for any ergodic process (Supplementary Fig. S3).
The anomality of the diffusion of MDC1 foci can also be recognized in the histogram of distance changes Δli for a single time interval Δt obtained for the carbon-irradiated samples (Fig. 3). Normal diffusion would lead to a Gaussian distribution of the probability density p(Δl, Δt) of the Δli(Δt) values for initial distances much larger than the diffusional length20. The measured distribution function differs significantly from a Gaussian distribution, but is well fitted by the distribution function for subdiffusion (adapted from20) when using an anomalous diffusion exponent α = 0.5 (see Fig. 3, where Δt = 60 s) and applying a two dimensional random walk situation for distance analysis and thus substituting x → Δl and Kα → 2dDα:

The differences to the Gaussian are visible both at very small distances, where a cusp-shaped histogram is obtained as predicted by the subdiffusion model (see plot on linear scale of Fig. 3a), as well as in the tails, where the distributions are significantly higher than predicted by a best Gaussian fit (log-plot of Fig. 3b). We conclude from our live-cell imaging and distance tracking over time periods between a few seconds and several hours after irradiation that IRIF do not diffuse normally but follow a subdiffusion process.
Histogram of distance changes (Δl(Δt = 60 s) between two neighbouring foci after carbon irradiation, fitted with the distribution function for normal and anomalous diffusion on linear (A) and logarithmic scale (B) (bin size = 0.03 μm, number of data points Ntotal = 5656).
SE-error bars are taken as  given through Poisson statistics.
given through Poisson statistics.
LET dependence of apparent foci mobility
To investigate whether IRIF diffusion depends on the density of damage sites along the irradiation track, the cells were also irradiated in a matrix of Δx = Δy = 6 μm with 32 protons (20 MeV at cell position, linear energy transfer LET = 2.6 keV/μm) per point. The number of protons was chosen to create on average 3 DSB per matrix point so that within the depth of view each detected IRIF contained on average only one DSB, while after carbon irradiation several DSB are expected to occur in each IRIF. As in the case of carbon-irradiated cells, a linear increase of log(σ2) with log(Δt) is evident (Fig. 4), demonstrating a power-law dependence of σ2 on Δt. For comparison, also the data obtained after carbon ion irradiation are shown.
Double-logarithmic plot of the squared standard deviations σ2 (±SEM) of the distance changes Δl(Δt) between neighbouring MDC1 foci in the nuclei of cells irradiated with carbon ions (filled squares, 657 foci pairs from 58 cells) and protons (open circles, 32 p per irradiation point, 99 foci pairs from 12 cells), each 30–60 time frames analyzed per sample.
The data are fitted with the power-law function (eq. (2)).
We determined α = 0.50 ± 0.05 (SEM) and D0.50 = (8.1 ± 1.8) × 10−4 μm2/s0.50 for the proton-irradiated cells, showing that the time dependence of foci after carbon and proton irradiation is very similar. As the anomality parameter α for proton and carbon irradiation (α = 0.50 ± 0.05 and α = 0.49 ± 0.05, respectively) is the same within the errors, the number of induced double-strand breaks within one focus does not seem to influence the degree of anomality of the foci diffusion. The absolute σ2 values, however, differ by a factor 2.2. This difference may be attributed to the fact that the dynamics observed in proton irradiated cells is mainly that of an isolated, single DSB (number of DSB per IRIF Np ~ 1), whereas due to clustered DSB induction and poor axial microscopic resolution a number of NC > 1 foci contribute to each detectable IRIF after carbon irradiation. Thus, the diffusional behaviour observed is that of the centre of mass of all DSB in this IRIF, where contributions of individual DSB can neutralize each other. In a pure statistical approach we expect σ2p/σ2C = NC/Np (see Supplementary Information “Random walk of the centre of mass of N DSB”). The measured ratio of 2.2 is approximately the number of DSB per micrometer as calculated in8 multiplied with the approximate depth of field of the used epifluorescence microscope.
To experimentally verify this assumption, we rationalized that increasing the microscopic resolution would allow to differentiate between the IRIF of individual DSB. With small-angle irradiation it is possible to visualize with good lateral resolution how ion-induced IRIF form along the track6. Due to technical limitation, we were not able to perform live-cell analysis after small-angle carbon irradiation. We decided, therefore, to reinvestigate data obtained in earlier work (Du et al. 201111) by NBS1 foci detection in carbon-irradiated cells fixed at various post-irradiation intervals. To allow a comparison of these data to our live-cell data, we introduce the “apparent diffusion coefficient” Dapp, which is defined as the ratio of the mean squared displacement MSD(Δt) (equation (1)) and the corresponding time Δt, corrected for the dimensionality d of the random walk:

Our live-cell analysis has d = 2 due to the two-dimensional analysis of the centre of foci in the focal plane of the epifluorescence microscope, while the analysis in11 has been performed in one dimension. The apparent diffusion coefficient Dapp is equal to the average diffusion constant that would result in the same MSD if normal diffusion had taken place. It is evident (Fig. 5) that the mobility of IRIF induced by small-angle carbon irradiation compares well to the mobility determined in the present work for proton-induced IRIF.
The apparent diffusion coefficient  of DSB induced by different ionizing radiation (A) and in comparison with that of centromeric, telomeric and interstitial chromosomal loci (B) (with standard errors).
of DSB induced by different ionizing radiation (A) and in comparison with that of centromeric, telomeric and interstitial chromosomal loci (B) (with standard errors).
C and p are the data sets from this work (cf. Fig. 4). (A) “C (Du)” is the lateral diffusion data of NBS1-foci in BJ1-hTERT cells irradiated with C ions (~40 MeV) at SNAKE11 and “Ni (Jakob)” and “γ (Krawczyk)” are the diffusion data of GFP-tagged 53BP1-foci in U2OS cells irradiated with 6 MeV Ni ions13 and with γ31, respectively. (B) Telomere data extracted from Bronstein et al25., Jegou et al19., De Vos et al33. and Krawczyk et al31. Apparent diffusion coefficient of centromeric and interstitial chromosomal loci calculated from Chubb et al18. and Krawczyk et al31. Dapp is calculated using eq. (4) with corresponding dimension d and calculating MSD according to eq. (1) in case of distance measurements.
Discussion
In the present work, we demonstrate that radiation-induced MDC1 foci, which are assumed to reflect the chromatin region surrounding one DSB or several clustered DSB, exhibit a subdiffusive mobility in the time range 5 s ≤ Δt ≤ 104 s, with α ≈ 0.5. Subdiffusive behaviour of chromatin regions has been described in yeast22, E. coli chromosomes23,24 and, at least in the time scale of up to 100 s, telomeric regions in mammalian U2OS cells25,26. Others, however, reported constrained diffusion of genomic loci (reviewed by21). Recently, Albert et al. (2012) proposed that genomic loci in general undergo subdiffusion at all time scales explored and that this effect may be overlooked when plotting MSD on a linear scale, where constrained and subdiffusive mobility lead to very similar curves17.
The underlying mechanisms leading to subdiffusion have mainly been investigated in the context of anomalous mobility of macromolecules such as proteins and microinjected beads in nucleoplasm and cytoplasm (e.g.27,28) where subdiffusion appears to be a consequence of crowding-induced viscoelasticity15,27. Modelling results suggest that subdiffusive motion of chromosomal loci in E. coli is a consequence of both viscoelasticity of the nucleoplasm and the restrictions on monomer mobility caused by neighbouring monomers in a polymer23. Also transient immobilization by binding to obstacles may contribute to subdiffusive behaviour29,30.
The most pertinent characteristic of subdiffusion is that apparent mobility becomes slower at longer observation times. To detect subdiffusive behaviour it is therefore important to avoid overestimation of mobility at long observation intervals which may be caused by insufficient correction for mobility of the reference system, i.e. the nucleus in the case of chromosomal loci. We derived Dapp from a variety of data on mobility measurements of chromosomal loci available in the literature (Fig. 5a + b.). We note that those few mobility measurements which did not show a reduction of Dapp with observation time were performed with single particle tracking (Fig. 5a “Ni (Jakob)”13, Fig. 5b “telomere (Bronstein)”25 at long Δt). In contrast, distance tracking as used in our work, intrinsically corrects for translational and rotational movements and, when using the standard deviation of distance changes as it is used here, even largely corrects for a change of the nucleus size. It is important to note that reduced apparent mobility at longer observation periods is not a consequence of altered conditions with increasing post-irradiation time (e.g. due to repair, or unphysiological conditions) since the mobility at short observation intervals is similar whether investigated within the first minutes after irradiation or after one hour (see Supplementary Fig. S3).
What are the consequences of subdiffusive mobility? As discussed by Weber (2010), “subdiffusive molecules take longer than expected to reach distant targets but they are also more likely to retrace local regions of space than a freely diffusing particle”23. Thus, the encountered subdiffusion predicts long-range movements of chromosomal loci and DSB to be rare, even at long time intervals. Figure 5a shows that our measurements for MDC1 and NBS1 foci11 mobility compare well with data on 53BP1 foci determined recently after γ irradiation31. Since contradictory data were published on whether IRIF can travel μm distances and coalesce to form repair centres7,12,13,32, it is interesting to compare IRIF mobility to that of undamaged chromatin regions. As evident from Fig. 5b, these data agree also well with a large set of published data on mobility of telomeric, centromeric, or interstitial chromosomal loci. In spite of the use of different methods and experimental systems, all available mobility data are surprisingly similar. Whether, as suggested by Krawczyk et al31., IRIF and telomeric loci exhibit generally a slightly enhanced mobility as compared to interstitial loci remains to be investigated in detail, also taking into account differences in foci substructures.
In any case, we conclude that any model invoking movements of foci over several μm within a few hours cannot be explained by typical mobility of chromosomal loci. Since the average distance of travel is small, the probability of joining wrong ends from DSB that initially were separated by several micrometers is low. For example, with typical values for Dapp smaller than 10−5 μm2/s at Δt = 3 h (Fig. 5b), the average sampled volume has a radius of  (d = 3) which is less than 0.5% of the total volume of the cell nucleus of about 700 μm3.
(d = 3) which is less than 0.5% of the total volume of the cell nucleus of about 700 μm3.
Our study investigated the mobility of IRIF surrounding break sites rather than the mobility of individual DNA ends. Little is known on the extent of tethering of both ends of a DSB site, resulting either from the original chromatin structure or from fast DSB detection and response mechanisms32. There must be a certain probability of separation between ends, since otherwise mis-rejoining and generation of aberrations would not occur. For our considerations, we assume that the single ends underlie subdiffusion of similar speed as the DSB site as a whole. If there was independent movement of both single ends at considerably faster velocity, the monitored IRIF would be expected to frequently dissociate into two foci, which was not seen, although rare dissociation events cannot be excluded from the limited statistics of the experiment. Furthermore, the DNA ends are still connected to the chromatin, which is found to follow subdiffusion also in the case of telomeric ends, with a mobility that agrees well with the IRIF mobility (cf. Fig. 5b). Computer simulations demonstrated that subdiffusion enhances the probability of finding nearby targets as compared to normal diffusion for initial separations R smaller than the so-called crossover radius Rc, thus facilitating cellular processes such as protein complex formation with high probability16. A target size a = 2 nm to find a broken DNA end approximated by the diameter of the size of the DNA end, a nucleoplasmic viscosity of 2 × 10−3 Pa·s at 37°C and an anomalous diffusion coefficient α = 0.5 lead to a search time-dependent crossover radius Rc [μm] ≈ 0.5 × tsearch1/4 (with tsearch in [s]) of more than 5 μm for search times of 1–3 hours (calculated according to16). The radius may be even enhanced by a factor 21/4 = 1.2 because both ends of a DSB are subject to subdiffusion. Provided that the distance of the two ends of a DSB that had separated from each other after damage induction is smaller than Rc, the probability that the ends encounter again would thus be larger in the case of subdiffusion than for normal diffusion. This means that subdiffusion supports the rejoining of ends during repair of DNA double-strand breaks. By labelling both sides of an enzyme-induced break site with different fluorescent molecules, Soutoglou et al. 2007 demonstrated typical separation distances in the range of R = 100 nm32, i.e. much smaller than the crossover radius Rc of more than 5 μm estimated here for several hours of search/repair time. Thus, the rejoining probability (within a few hours after irradiation) can be estimated to be larger than 90% for subdiffusion (using the model of16), while normal diffusion would only rejoin broken ends with initial separation R ~ 100 nm with maximum 2% probability (P(R) = a/R16).
When there are more than one DSB within the initial separation distance R of the two ends of a single DSB, the probability to connect two ends would be the same for all DNA ends, thus resulting in a large fraction of translocations. Note that the ratio of correctly rejoined ends and translocations would be the same under subdiffusion or normal diffusion random walk, but many more single ends would be left in case of a normal diffusion. In the case that two DSB are created at a distance larger than R, however, the ratio of correctly rejoined ends to translocations increases more for subdiffusion than for normal diffusion.
In conclusion, we propose that the subdiffusional behaviour of DSB sites guarantees a high probability of rejoining the ends of an individual DSB while reducing the probability of chromosomal aberrations produced by rejoining the ends of different DSB or of leaving a large fraction of DNA ends unrepaired.
Methods
Cell culture and irradiation
Generation of a stable transfectant (clone F1) of human osteosarcoma cells (U2OS) with pEGFP-MDC134 was seeded into custom-made cell containers35 in HEPES-buffered, phenol red-free medium supplemented with 0.25 mM Trolox at 37°C and 5% CO2 (72 hours before irradiation).
The irradiation was performed at the microbeam facility SNAKE36,37 at the Munich 14 MV tandem accelerator with one carbon ion (43 MeV, LET = 370 keV/μm) or 32 protons (20 MeV, LET = 2.6 keV/μm) per point in a 5 × 5 μm2 matrix, with an accuracy of 0.58 μm in x and 0.86 μm in y35 (see Fig. 1). One single 43 MeV carbon ion results in approximately 20 DSB along its approximately 7 μm track through the nucleus8 and the lowest number of protons necessary to obtain observable foci, which was about 30 protons per point, means an average number of three DSB that are distributed randomly along the ion track across the cell nucleus that has a height of about (7.2 ± 0.3) μm. During irradiation and the subsequent observation the cells were covered by cell culture medium and the temperature was kept constant at 37°C.
Distance tracking with 2D Fluorescence Microscopy and quantitative dynamics analysis
Live cell observation started approximately 200–400 s after irradiation and time series of different interval length Δt (e.g. 5 s, 1 or 10 minutes) and total length ttotal (20 minutes up to several hours) were recorded. Motion analysis was performed with the open source image analysis software ImageJ (http://rsb.info.nih.gov/ij) using the SpotTracker plugin (http://bigwww.ep.ch/sage/soft/spottracker/), developed by Daniel Sage et al38. for tracking fluorescent particles in dynamic image sequences with subpixel resolution. After visually verifying that the foci were tracked correctly, the two-dimensional distance l(t) of all neighboring foci pairs (as well as second next neighbors) were calculated from the x-y coordinates gathered by the SpotTracker algorithm and the standard deviation σ of the distance changes Δli was determined for different time intervals Δt for analysis of the foci dynamics. For more details see Supplementary Information.
References
Hlatky, L. Double-strand break motions shift radiation risk notions? Proc. Natl. Acad. Sci. U.S.A. 109, 351–352 (2012).
Savage, J. R. K. Insight into sites. Mutat. Res. 366, 81–95 (1996).
Savage, J. R. K. Reflections and meditations upon complex chromosomal exchanges. Mutat. Res. 512, 93–109 (2002).
Sachs, R. K. Chen, A. M. & Brenner, D. J. Review: proximity effects in the production of chromosome aberrations by ionizing radiation. Int. J. Radiat. Biol. 71, 1–19 (1997).
Friedland, W. Jacob, P. & Kundrat, P. Stochastic simulation of DNA double-strand break repair by non-homologous end joining based on track structure calculations. Radiat Res 173, 677–688 (2010).
Jakob, B. Scholz, M. & Taucher-Scholz, G. Biological imaging of heavy charged-particle tracks. Radiat. Res. 159, 676–684 (2003).
Aten, J. A. et al. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303, 92–95 (2004).
Hauptner, A. et al. Spatial Distribution of DNA Double-Strand Breaks from Ion Tracks. Royal Danish Academy of Sciences and Letters, Copenhagen 59–85 (2006b).
Hable, V. et al. Methods for quantitative evaluation of dynamics of repair proteins within irradiated cells. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms 245, 298–301 (2006).
Costes, S. V. et al. Image-based modeling reveals dynamic redistribution of DNA damage into nuclear sub-domains. PLoS Comput. Biol. 3, e155 (2007).
Du, G. et al. Spatial dynamics of DNA damage response protein foci along the ion trajectory of high-LET particles. Radiat. Res. 176, 706–715 (2011).
Neumaier, T. et al. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proc. Natl. Acad. Sci. U.S.A. 109, 443–448 (2012).
Jakob, B. Splinter, J. Durante, M. & Taucher-Scholz, G. Live cell microscopy analysis of radiation-induced DNA double-strand break motion. Proceedings of the National Academy of Sciences 106, 3172–3177 (2009).
Kruhlak, M. J. et al. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 172, 823–834 (2006).
Guigas, G. Kalla, C. & Weiss, M. Probing the nanoscale viscoelasticity of intracellular fluids in living cells. Biophys. J. 93, 316–323 (2007).
Guigas, G. & Weiss, M. Sampling the cell with anomalous diffusion - the discovery of slowness. Biophys. J. 94, 90–94 (2008).
Albert, B., Léger-Silvestre, I., Normand, C. & Gadal, O. Nuclear organization and chromatin dynamics in yeast: Biophysical models or biologically driven interactions? Biochim. Biophys. Acta 1819, 468–481 (2012).
Chubb, J. R. Boyle, S. Perry, P. & Bickmore, W. A. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12, 439–445 (2002).
Jegou, T. et al. Dynamics of telomeres and promyelocytic leukemia nuclear bodies in a telomerase-negative human cell line. Mol. Biol. Cell 20, 2070–2082 (2009).
Metzler, R. & Klafter, J. The random walk's guide to anomalous diffusion: a fractional dynamics approach. Physics Reports 339, 1–77 (2000).
Lanctôt, C. et al. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 8, 104–115.
Cabal, G. G. et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441, 770–773 (2006).
Weber, S. C. Spakowitz, A. J. & Theriot, J. A. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys. Rev. Lett. 104, 238102 (2010).
Weber, S. C. Spakowitz, A. J. & Theriot, J. A. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc. Natl. Acad. Sci. U.S.A. 109, 7338–7343 (2012).
Bronstein, I. et al. Transient anomalous diffusion of telomeres in the nucleus of mammalian cells. Phys. Rev. Lett. 103, 18102 (2009).
Kepten, E. Bronshtein, I. & Garini, Y. Improved estimation of anomalous diffusion exponents in single-particle tracking experiments. Phys. Rev. E 87 (2013).
Weiss, M. Elsner, M. Kartberg, F. & Nilsson, T. Anomalous subdiffusion is a measure for cytoplasmic crowding in living cells. Biophys. J. 87, 3518–3524 (2004).
Wachsmuth, M. Waldeck, W. & Langowski, J. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol. 298, 677–689 (2000).
Banks, D. S. & Fradin, C. Anomalous diffusion of proteins due to molecular crowding. Biophys. J. 89, 2960–2971 (2005).
Lukacs, G. L. et al. Size-dependent DNA mobility in cytoplasm and nucleus. J. Biol. Chem. 275, 1625–1629 (2000).
Krawczyk, P. M. et al. Chromatin mobility is increased at sites of DNA double-strand breaks. J. Cell. Sci. 125, 2127–2133 (2012).
Soutoglou, E. et al. Positional stability of single double-strand breaks in mammalian cells. Nat. Cell Biol. 9, 675–682 (2007).
de Vos, W. H. et al. Four-dimensional telomere analysis in recordings of living human cells acquired with controlled light exposure microscopy. J Microsc 238, 254–264 (2010).
Hable, V. et al. Recruitment Kinetics of DNA repair proteins Mdc1 and Rad52 but not 53BP1 depend on damage complexity. PLoS ONE 7, e41943 (2012).
Hable, V. et al. The live cell irradiation and observation setup at SNAKE. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms 267, 2090–2097 (2009).
Datzmann, G. et al. The Munich microprobe SNAKE: First results using 20 MeV protons and 90 MeV sulfur ions. 7th International Conference on Nuclear Microprobe Technology and Applications. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms 181, 20–26 (2001).
Hauptner, A. et al. Microirradiation of cells with energetic heavy ions. Radiat Environ Biophys 42, 237–245 (2004).
Sage, D. Neumann, F. R. Hediger, F. Gasser, S. M. & Unser, M. Automatic tracking of individual fluorescence particles: application to the study of chromosome dynamics. IEEE Trans Image Process 14, 1372–1383 (2005).
Acknowledgements
The authors acknowledge the financial support of the DFG Cluster of Excellence: Munich-Centre for Advanced Photonics (MAP), the German Federal Ministry for the Environment, Nature Conservation and Nuclear Safety (BMU) (Project No. 3610S30015) and by the Maier Leibnitz Laboratory Munich.
Author information
Authors and Affiliations
Contributions
S.G., V.H. and G.D. designed research; S.G., V.H., C.G., G.A.D. and C.S. performed research; S.G., V.H., M.H. and G.D. analyzed data; and S.G., V.H., G.D. and A.A.F. wrote the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/
About this article
Cite this article
Girst, S., Hable, V., Drexler, G. et al. Subdiffusion Supports Joining Of Correct Ends During Repair Of DNA Double-Strand Breaks. Sci Rep 3, 2511 (2013). https://doi.org/10.1038/srep02511
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02511
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.